
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,083,496 – 16,083,627 |
| Length | 131 |
| Max. P | 0.964737 |

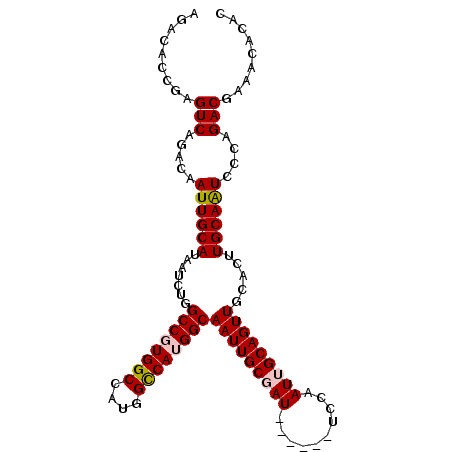
| Location | 16,083,496 – 16,083,601 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -29.19 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
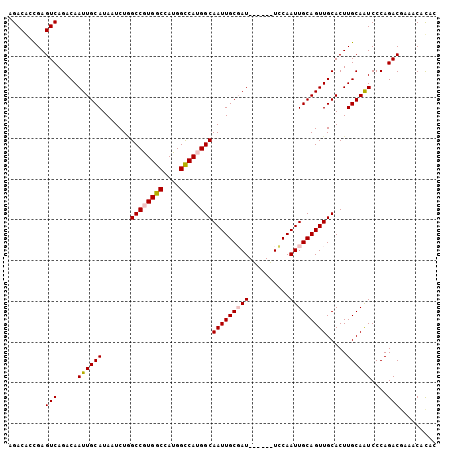
>2R_DroMel_CAF1 16083496 105 - 20766785 AGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAAUUGCAGUUGCACUUGCAAUCCCAGACGAGACACAC .(((.....)))..............(((((.((((((....))))))((((.((((((((((((....)))))))))))).))))....))))).......... ( -40.50) >DroPse_CAF1 8347 99 - 1 AGACAUUGAGUCAGACAAUUGCAUAAUCUGGCCAUGGCCAUGGCCACGGCAAUUGCCAU------UGCAAUUGCAGUUGCACUUGCAGUCCCAGACACAACAUGG .....(((.(((.(.(((((((......(((((((....)))))))..))))))).)((------(((((.(((....))).)))))))....))).)))..... ( -34.20) >DroEre_CAF1 5151 99 - 1 CGACGCCGAGUCAGACAAUUGCAUAAUCUUGCCGUGGCCAUGGUCAUGGCAAUUGCGAU------UGCAAUUGCAGUUGCACUUGCAAUCCCAGACGAAGCACAC ....((...(((.....((((((.....((((((((((....)))))))))).((((((------(((....)))))))))..))))))....)))...)).... ( -39.00) >DroYak_CAF1 5250 99 - 1 AGACGCCGAGUCAGACAAUUGCAUAAUCUAGCCGUGGCCAUGGCCAUGGCAAUUGCGAU------UCCAAUUGCAGUUGCACUUGCAAUCCCAGACGAAACACAC .(((.....))).(.(((.(((........((((((((....))))))))(((((((((------....)))))))))))).))))................... ( -33.40) >DroAna_CAF1 5398 105 - 1 GGACAUUGAGUCAGACAAUUGCAUAAUCUAGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUACGAUUCCAAUUGCAGUUGCACUUGCAAUCCCAGACUUGCCUCGC ((.....(((((.....((((((.......((((((((....))))))))((((((((((........)))))))))).....))))))....))))).)).... ( -36.40) >DroPer_CAF1 8310 99 - 1 AGACAUUGAGUCAGACAAUUGCAUAAUCUGGCCAUGGCCAUGGCCACGGCAAUUGCCAU------UGCAAUUGCAGUUGCACUUGCAGUCCCAGACACAACAUGG .....(((.(((.(.(((((((......(((((((....)))))))..))))))).)((------(((((.(((....))).)))))))....))).)))..... ( -34.20) >consensus AGACACCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAU______UCCAAUUGCAGUUGCACUUGCAAUCCCAGACGAAACACAC .........(((.....((((((.......((((((((....))))))))(((((((((..........))))))))).....))))))....)))......... (-29.19 = -29.50 + 0.31)

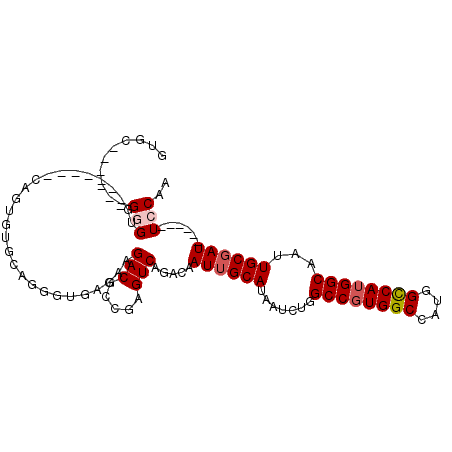
| Location | 16,083,531 – 16,083,627 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
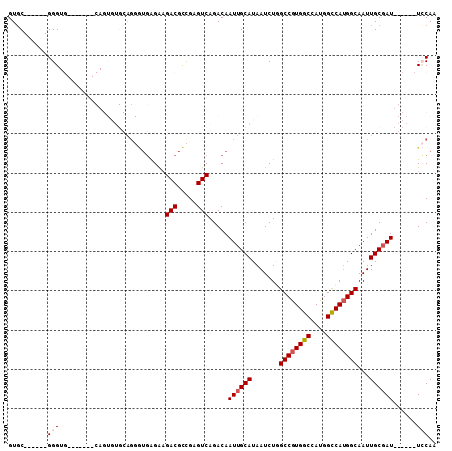
>2R_DroMel_CAF1 16083531 96 - 20766785 GUGC------GGGUG-------CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAA .(((------(.(..-------...).)))).((((......))))(((((.(.((((((((.......((((((((....))))))))...))))))))))))))... ( -37.80) >DroPse_CAF1 8382 79 - 1 GUCC------GGG-----------------AG-ACCGGGAGACAUUGAGUCAGACAAUUGCAUAAUCUGGCCAUGGCCAUGGCCACGGCAAUUGCCAU------UGCAA .(((------((.-----------------..-.))))).(((.....)))......(((((.....(((((((....))))))).(((....)))..------))))) ( -33.10) >DroSec_CAF1 5139 96 - 1 GUGC------GGGUG-------CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAA .(((------(.(..-------...).)))).((((......))))(((((.(.((((((((.......((((((((....))))))))...))))))))))))))... ( -37.80) >DroSim_CAF1 5273 102 - 1 GUGCGGGUGCGGGUG-------CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAA .((((.((((....)-------)).).)))).((((......))))(((((.(.((((((((.......((((((((....))))))))...))))))))))))))... ( -41.60) >DroEre_CAF1 5186 76 - 1 GUGC------GGG---------------------UGGGACGACGCCGAGUCAGACAAUUGCAUAAUCUUGCCGUGGCCAUGGUCAUGGCAAUUGCGAU------UGCAA ....------.((---------------------((......))))......(.((((((((.....((((((((((....)))))))))).))))))------))).. ( -30.90) >DroYak_CAF1 5285 97 - 1 GUGC------GGGUGCAGUGUGCAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUAGCCGUGGCCAUGGCCAUGGCAAUUGCGAU------UCCAA ....------((((((((((((((((((((..((((......))))..))...)).)))))))......((((((((....)))))))).))))))..------))).. ( -37.40) >consensus GUGC______GGGUG_______CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAU______UCCAA ..........(((...........................(((.....))).....((((((.......((((((((....))))))))...))))))......))).. (-22.44 = -22.97 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:23 2006