| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,079,546 – 16,079,638 |
| Length | 92 |
| Max. P | 0.795218 |

| Location | 16,079,546 – 16,079,638 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
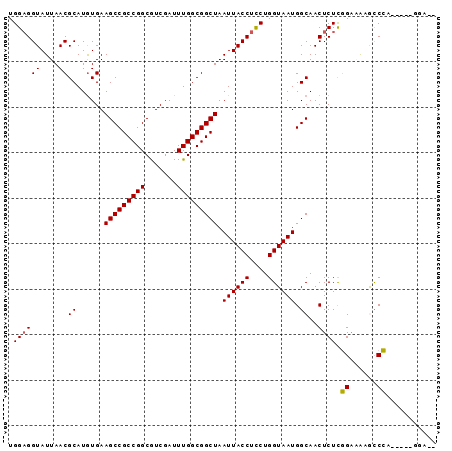
>2R_DroMel_CAF1 16079546 92 + 20766785 UGGAGGUAUUAACGCAUGUGAAGCCGCCGGCGUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUCGGAAAGGCCCAAGAAAGGA-- .(((((((....((....)).(((((((((.......)))))))))...))))))).......(....)(((.((......)).))).....-- ( -28.70) >DroPse_CAF1 4079 86 + 1 UGGAGGUAUUAACGCAUGUGAAGCCGCCGGCAUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUCCGGUGUACCG------GUA-- .(((((((....((....)).(((((((((.......)))))))))...))))))).......(....)...((((....)))------)..-- ( -31.50) >DroSim_CAF1 1173 92 + 1 UGGAGGUAUUAACGCAUGUGAAGCCGCCGGCGUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUCGGAAAGGCCCAAGAAAGGA-- .(((((((....((....)).(((((((((.......)))))))))...))))))).......(....)(((.((......)).))).....-- ( -28.70) >DroEre_CAF1 1213 93 + 1 UGG-GGUAUUAACGCAUGUGAAGCCGCCGGCGUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUUGGAAAAGUCCAAGAAUGGAGA ...-.........((......(((((((((.......))))))))).((((((....)))))).))...(((((((....)))))))....... ( -31.10) >DroAna_CAF1 1144 88 + 1 UGGAGGUAUUAACGCAUAUGAAGCCGCCGGCGUCGAUUUGGCGGCUAAUUACCUUCUGGUAAUGGCAACUCUCGGAAAAGCCCG------GAGA .............((......(((((((((.......))))))))).((((((....)))))).))..(((.(((......)))------))). ( -30.20) >DroPer_CAF1 4035 86 + 1 UGGAGGUAUUAACGCAUGUGAAGCCGCCGGCAUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUCCGGUGUACCG------GUA-- .(((((((....((....)).(((((((((.......)))))))))...))))))).......(....)...((((....)))------)..-- ( -31.50) >consensus UGGAGGUAUUAACGCAUGUGAAGCCGCCGGCGUCGAUUUGGCGGCUAAUUACCUCCUGGUAAUGGCAACUCUCGGAAAAGCCCA_____GGA__ .((((((....))((......(((((((((.......))))))))).((((((....)))))).))..)))).((......))........... (-25.78 = -25.50 + -0.28)

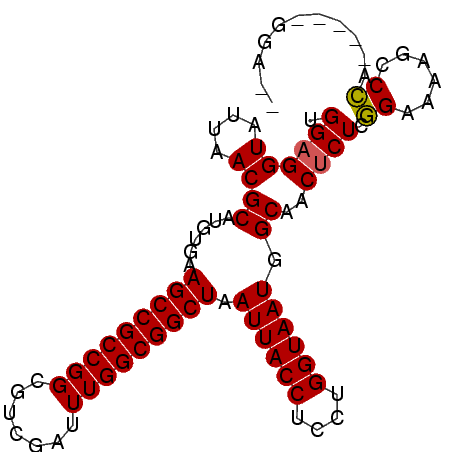
| Location | 16,079,546 – 16,079,638 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -23.71 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
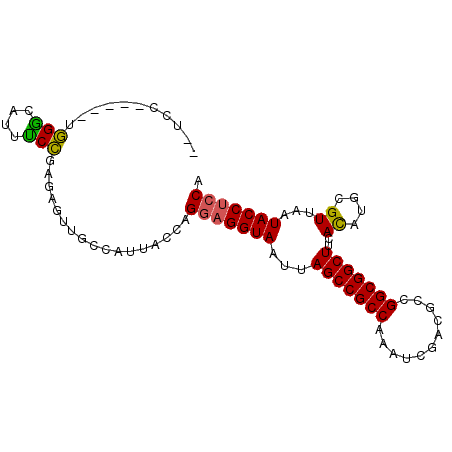
>2R_DroMel_CAF1 16079546 92 - 20766785 --UCCUUUCUUGGGCCUUUCCGAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGACGCCGGCGGCUUCACAUGCGUUAAUACCUCCA --....((((((((....))))))))............(((((((...(((((((...........)))))))..((....))...))))))). ( -30.60) >DroPse_CAF1 4079 86 - 1 --UAC------CGGUACACCGGAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGAUGCCGGCGGCUUCACAUGCGUUAAUACCUCCA --..(------(((....))))................(((((((...(((((((...........)))))))..((....))...))))))). ( -30.00) >DroSim_CAF1 1173 92 - 1 --UCCUUUCUUGGGCCUUUCCGAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGACGCCGGCGGCUUCACAUGCGUUAAUACCUCCA --....((((((((....))))))))............(((((((...(((((((...........)))))))..((....))...))))))). ( -30.60) >DroEre_CAF1 1213 93 - 1 UCUCCAUUCUUGGACUUUUCCAAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGACGCCGGCGGCUUCACAUGCGUUAAUACC-CCA .....(((((((((....))))))))).((.((((((....)))))).(((((((...........)))))))......)).........-... ( -28.20) >DroAna_CAF1 1144 88 - 1 UCUC------CGGGCUUUUCCGAGAGUUGCCAUUACCAGAAGGUAAUUAGCCGCCAAAUCGACGCCGGCGGCUUCAUAUGCGUUAAUACCUCCA .(((------((((....)))).)))..((.((((((....)))))).(((((((...........)))))))......))............. ( -25.00) >DroPer_CAF1 4035 86 - 1 --UAC------CGGUACACCGGAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGAUGCCGGCGGCUUCACAUGCGUUAAUACCUCCA --..(------(((....))))................(((((((...(((((((...........)))))))..((....))...))))))). ( -30.00) >consensus __UCC_____UGGGCAUUUCCGAGAGUUGCCAUUACCAGGAGGUAAUUAGCCGCCAAAUCGACGCCGGCGGCUUCACAUGCGUUAAUACCUCCA ...........(((....))).................(((((((...(((((((...........)))))))..((....))...))))))). (-23.71 = -23.10 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:21 2006