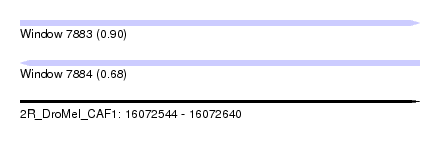
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,072,544 – 16,072,640 |
| Length | 96 |
| Max. P | 0.895007 |

| Location | 16,072,544 – 16,072,640 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -19.06 |
| Energy contribution | -20.56 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.895007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16072544 96 + 20766785 AUAAGCCAUCCAU-UCCAUAUCCACAUUUUCGUCGA----------CACCAACGCAGUCGACUAAUGAUGCAAAUGUCACAAGCGAACAAAGAUCGCUGGCGAUCUC ....(((......-..(((...((((((...(((((----------(.........)))))).)))).))...))).....(((((.......))))))))...... ( -20.90) >DroSec_CAF1 693 100 + 1 AUACGCCAUCCAU-CCCAUAUCCACAUCUUCGUCGACCG------CCACCAACGCAGUCGACUAAUGAUGCGGAUGUCACAAGCGAACAAAGAACGCUGGCGAUCUC ...((((......-...((((((.((((((.((((((.(------(.......)).)))))).)).)))).))))))....((((.........))))))))..... ( -33.70) >DroSim_CAF1 690 100 + 1 AUACGCCAUCCAU-CCCAUAUCCACAUCUACGUCGACCG------CCACCAACGCAGUCGACUAAUGAUGCGGAUGUCACAAGCGAACAAAGAACGCUGGCGAUCUC ...((((......-...((((((.((((...((((((.(------(.......)).))))))....)))).))))))....((((.........))))))))..... ( -33.60) >DroEre_CAF1 774 107 + 1 AUGUGCCAUCCAUAGCCAUAUCCACAUCCUCGUCCACCGCCACCGCCACCAACGCAGUCGACUAAUGAUGCAAGUGUCACAAGCGGACAAAGAGCGGUGGCGAUCUC (((.((........)))))..................((((((((((......(((.(((.....))))))...((((.......))))..).)))))))))..... ( -27.90) >consensus AUACGCCAUCCAU_CCCAUAUCCACAUCUUCGUCGACCG______CCACCAACGCAGUCGACUAAUGAUGCAAAUGUCACAAGCGAACAAAGAACGCUGGCGAUCUC ...((((..........((((((.((((...((((((...................))))))....)))).))))))....((((.........))))))))..... (-19.06 = -20.56 + 1.50)

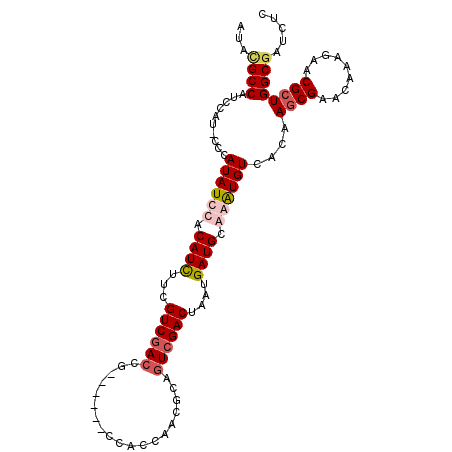

| Location | 16,072,544 – 16,072,640 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.26 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16072544 96 - 20766785 GAGAUCGCCAGCGAUCUUUGUUCGCUUGUGACAUUUGCAUCAUUAGUCGACUGCGUUGGUG----------UCGACGAAAAUGUGGAUAUGGA-AUGGAUGGCUUAU ......((((((((.......))))(..(..(((.(.((.((((.((((((.((....)))----------)))))...)))))).).)))..-)..).)))).... ( -27.60) >DroSec_CAF1 693 100 - 1 GAGAUCGCCAGCGUUCUUUGUUCGCUUGUGACAUCCGCAUCAUUAGUCGACUGCGUUGGUGG------CGGUCGACGAAGAUGUGGAUAUGGG-AUGGAUGGCGUAU .....(((((.((((((....(((....))).(((((((((....(((((((((.......)------))))))))...)))))))))..)))-)))..)))))... ( -42.50) >DroSim_CAF1 690 100 - 1 GAGAUCGCCAGCGUUCUUUGUUCGCUUGUGACAUCCGCAUCAUUAGUCGACUGCGUUGGUGG------CGGUCGACGUAGAUGUGGAUAUGGG-AUGGAUGGCGUAU .....(((((.((((((....(((....))).(((((((((....(((((((((.......)------))))))))...)))))))))..)))-)))..)))))... ( -42.50) >DroEre_CAF1 774 107 - 1 GAGAUCGCCACCGCUCUUUGUCCGCUUGUGACACUUGCAUCAUUAGUCGACUGCGUUGGUGGCGGUGGCGGUGGACGAGGAUGUGGAUAUGGCUAUGGAUGGCACAU ......((((((((((((((((((((.((((........))))..(((.(((((.......)))))))))))))))))))).))))...)))).............. ( -38.30) >consensus GAGAUCGCCAGCGUUCUUUGUUCGCUUGUGACAUCCGCAUCAUUAGUCGACUGCGUUGGUGG______CGGUCGACGAAGAUGUGGAUAUGGG_AUGGAUGGCGUAU ......(((((((.........))).......(((((((((....((((((((...............))))))))...)))))))))...........)))).... (-24.01 = -25.26 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:17 2006