| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,070,789 – 16,070,895 |
| Length | 106 |
| Max. P | 0.877842 |

| Location | 16,070,789 – 16,070,895 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
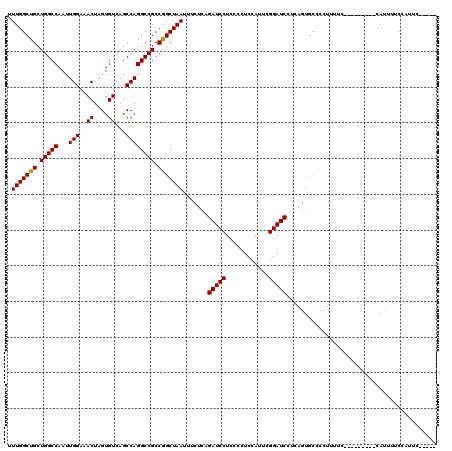
>2R_DroMel_CAF1 16070789 106 + 20766785 UUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCGGCUAAUUUCUCAGAUCCUCCCCUCCAUUCGGAUCCUGAGUGCCCCUUUUC---------CAUUUUCCAUUC----- ...((((((((((.....(....)....))))))..))))...(((......(((((((((............)))).))))).))).......---------............----- ( -29.70) >DroSec_CAF1 124532 90 + 1 UUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCGGCUAAUUUCUCAGAUCCUCCCCUCCAUUCGGAUC-------------------------CAUUUUCCAUUC----- .(((((((.(((((...(((..((....))...)))))))).))))))).......(((((............)))))-------------------------............----- ( -24.80) >DroSim_CAF1 131332 106 + 1 UUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCGGCUAAUUUCUCAGAUCCUCCCCUCCAUUCGGAUCCUGAGUGCCCCUUUUC---------CAUUUUCCAUUC----- ...((((((((((.....(....)....))))))..))))...(((......(((((((((............)))).))))).))).......---------............----- ( -29.70) >DroEre_CAF1 131425 120 + 1 GUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCGGCUAAUUUCUCAGAUCCUCCCCUCCAUUCGGAUCCGCAGUGCCCCUUUCCACAUUUCCACAUUUCCCAUACCCCGA ((((((((.(((((...(((..((....))...)))))))).))))))))......(((((............)))))....(((........)))........................ ( -26.80) >DroYak_CAF1 136019 103 + 1 GUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCAGCUAAUUUCUCAGAUCCUCCCCUCCAUUGGGAUCCUCAGUGCCCCUUUUC---------CAUUUCCC--------A ((((((((.(((((...(((..((....))...)))))))).))))))))......((((((..........))))))................---------........--------. ( -27.10) >consensus UUUGGCUGCUGGCCAAUUGGAAACUAGUGUCAGCCAGGCCGCCGGCUAAUUUCUCAGAUCCUCCCCUCCAUUCGGAUCCUCAGUGCCCCUUUUC_________CAUUUUCCAUUC_____ .(((((((.(((((...(((..((....))...)))))))).))))))).......(((((............))))).......................................... (-25.02 = -24.86 + -0.16)

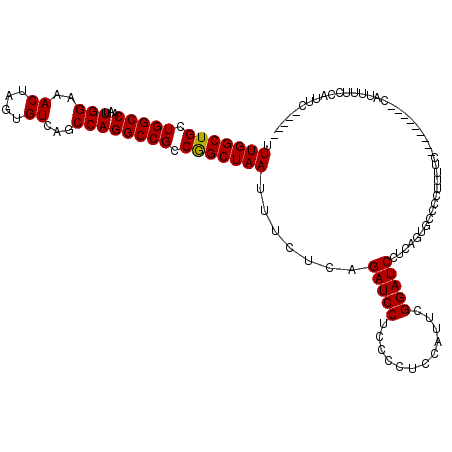
| Location | 16,070,789 – 16,070,895 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
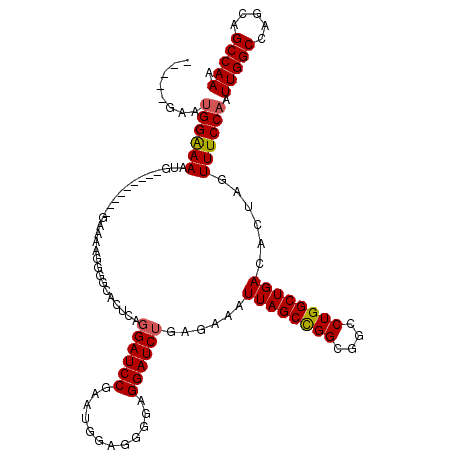
>2R_DroMel_CAF1 16070789 106 - 20766785 -----GAAUGGAAAAUG---------GAAAAGGGGCACUCAGGAUCCGAAUGGAGGGGAGGAUCUGAGAAAUUAGCCGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAA -----..........((---------((((..((...(((((.((((............)))))))))...((((((((....))))))))..))..)))))).(((((.....))))). ( -35.90) >DroSec_CAF1 124532 90 - 1 -----GAAUGGAAAAUG-------------------------GAUCCGAAUGGAGGGGAGGAUCUGAGAAAUUAGCCGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAA -----...((((((..(-------------------------(((((............))))))......((((((((....))))))))......)))))).(((((.....))))). ( -29.00) >DroSim_CAF1 131332 106 - 1 -----GAAUGGAAAAUG---------GAAAAGGGGCACUCAGGAUCCGAAUGGAGGGGAGGAUCUGAGAAAUUAGCCGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAA -----..........((---------((((..((...(((((.((((............)))))))))...((((((((....))))))))..))..)))))).(((((.....))))). ( -35.90) >DroEre_CAF1 131425 120 - 1 UCGGGGUAUGGGAAAUGUGGAAAUGUGGAAAGGGGCACUGCGGAUCCGAAUGGAGGGGAGGAUCUGAGAAAUUAGCCGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAC ....(((.(((..(((.((((((((((..........((.(((((((............)))))))))...((((((((....)))))))))))..))))))))))..)))...)))... ( -38.00) >DroYak_CAF1 136019 103 - 1 U--------GGGAAAUG---------GAAAAGGGGCACUGAGGAUCCCAAUGGAGGGGAGGAUCUGAGAAAUUAGCUGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAC .--------........---------.......(((.((.(((.((((.......))))...))).))......((((((.(..(((..(((....))).)))..).)))))).)))... ( -32.50) >consensus _____GAAUGGAAAAUG_________GAAAAGGGGCACUCAGGAUCCGAAUGGAGGGGAGGAUCUGAGAAAUUAGCCGGCGGCCUGGCUGACACUAGUUUCCAAUUGGCCAGCAGCCAAA ........((((((...........................((((((............))))))......((((((((....))))))))......)))))).(((((.....))))). (-26.70 = -26.70 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:14 2006