
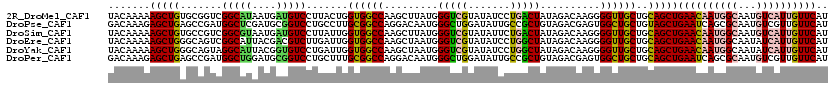
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,065,705 – 16,065,825 |
| Length | 120 |
| Max. P | 0.645912 |

| Location | 16,065,705 – 16,065,825 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
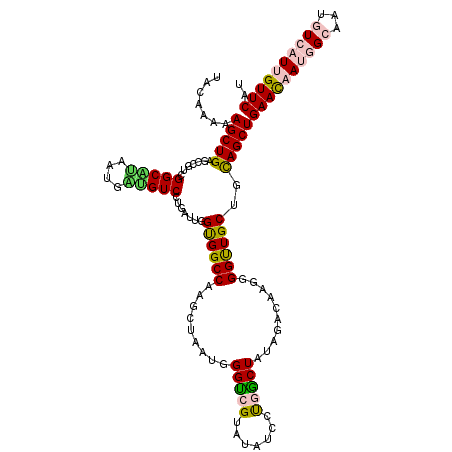
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16065705 120 + 20766785 UACAAAAAGCUGUGCGGUCGGCAUAAUGAUGUCCUUACUGGUGGCCAAGCUUAUGGGUCGUAUAUCCUGACUAUAGACAAGGGGUUGCUGCAGCUGAACAAUGGCAAUGUCAUUGUUCAU .......((((((((((((((.(((...(((.(((....(((......)))...))).))).))).))))))...(((.....))))).))))))((((((((((...)))))))))).. ( -37.50) >DroPse_CAF1 140585 120 + 1 GACAAAGAGCUGAGCCGAUGGCUCGAUGCGGUCCUGCCUUGCGGCCAGGACAAUGGGCUGGAUAUUGCCGCUGUAGACGAGUGGCUGCUGUAGCUGAAUCAGCGCAAUGUCGUUGUUCAU (((((....(((.((((..(((..(((...)))..)))...)))))))((((.((.(((((.....((((((.......)))))).((....))....))))).)).)))).)))))... ( -44.50) >DroSim_CAF1 123341 120 + 1 UACAAAAAGCUGUGCCGUCGGCGUAAUGAUGUCCUUAUUGGUGGCCAAGCUUAUGGGUCGUAUAUUCUGACUAUAGACAAGGGGUUGCUGCAGCUGAACAAUGGCAAUGUCAUUGUUCAU .......((((((((.(((((.(((...(((.(((....(((......)))...))).))).))).)))))....(((.....))))).))))))((((((((((...)))))))))).. ( -35.80) >DroEre_CAF1 126192 120 + 1 UACAAAAAGCUGGGCAGUCGGCAUUACGACGUCUUGAUUGGUGGCCAAGCUAAUGGGUCGUAUAUCCUGGCUAUAGACAAGGGGUUGCUGCAGCUGAACAAUGGCAAUAUCAUUGUUCAU .......(((((((((((((......)))).(((((.((.(((((((.......((((.....))))))))))).)))))))...)))).)))))(((((((((.....))))))))).. ( -40.61) >DroYak_CAF1 130732 120 + 1 UACAAAAAGCUGGGCAGUAGGCAUUACGGUGUCCUGAUUGGUGGCCAAGCUAAUGGGUCGUAUAUCCUGGCUAUAGACAAGGGGUUGCUGCAGCUGAACAAUGGCAAUAUCAUUGUUCAU .......((((..(((((((((((....))))(((.....(((((((.......((((.....))))))))))).....)))..)))))))))))(((((((((.....))))))))).. ( -36.71) >DroPer_CAF1 134250 120 + 1 GACAAAGAGCUGAGCCGAUGGCUGGAUGCGGUCCUGCUUUGCGGCCAGGACAAUGGGCUGGAUAUUGCCGCUGUAGACGAGUGGCUGCUGCAGCUGAAUCAGCGCAAUGUCGUUGUUCAU (((((....(((.((((..(((.((((...)))).)))...)))))))((((.((.(((((.....((((((.......)))))).((....))....))))).)).)))).)))))... ( -46.10) >consensus UACAAAAAGCUGAGCCGUCGGCAUAAUGAUGUCCUGAUUGGUGGCCAAGCUAAUGGGUCGUAUAUCCUGGCUAUAGACAAGGGGUUGCUGCAGCUGAACAAUGGCAAUGUCAUUGUUCAU .......(((((.......(((((....))))).......((((((.........(((((.......)))))..........))))))..)))))((((((((((...)))))))))).. (-27.41 = -27.24 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:06 2006