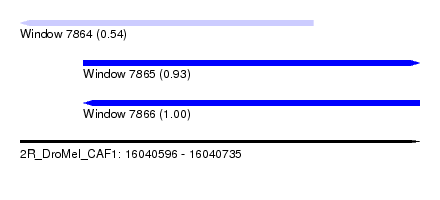
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,040,596 – 16,040,735 |
| Length | 139 |
| Max. P | 0.996800 |

| Location | 16,040,596 – 16,040,698 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16040596 102 - 20766785 UUGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUACAAACCUCGC (..(((.(((...(((...(((((((.........))))))).......(((....)))))).))))))..)(((((.....)))))............... ( -20.50) >DroSec_CAF1 95491 100 - 1 UCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUGCAA--UUCGC ((((((.(((...(((...(((((((.........))))))).......(((....)))))).)))))))))....(((.(((........))).--))).. ( -26.60) >DroSim_CAF1 98867 101 - 1 UCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUUCAA-CCUCGC ......(((.((.(((...(((((((.........))))))).......(((....)))))).))((((((((((((.....)))))....))))-)))))) ( -23.90) >DroEre_CAF1 102159 101 - 1 UCGGCCGCGUGCCCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUACAA-CCUCGC ..(((.....)))(((...(((((((.........))))))).......(((....)))))).((((((((.(((((.....))))).....)))-)).))) ( -21.90) >DroYak_CAF1 105699 101 - 1 UCGGCCGCGUGCCCCGAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUACAA-CCUCGC ..(((.....)))(((...(((((((.........))))))).......(((....)))))).((((((((.(((((.....))))).....)))-)).))) ( -21.20) >consensus UCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAGUGCUUUAAUCUACAA_CCUCGC ((((((.(((...(((...(((((((.........))))))).......(((....)))))).)))))))))(((((.....)))))............... (-21.92 = -21.36 + -0.56)

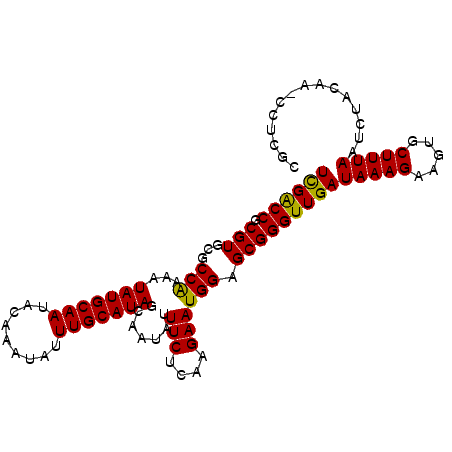
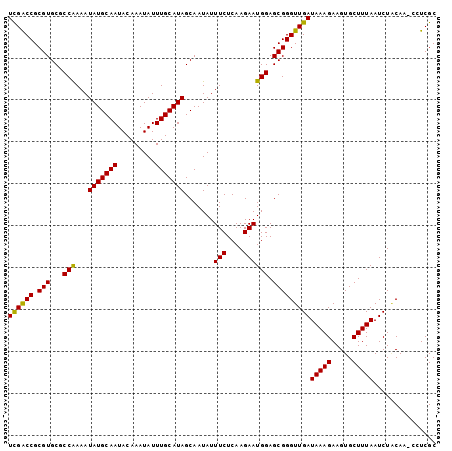
| Location | 16,040,618 – 16,040,735 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -29.83 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16040618 117 + 20766785 CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUUGGCGCACGCGGUCAAAUUCGCGAAUUGCAGGGGUAGAUAGCAAAGUGGUUUG (((..(((((.(((((((.((....)).(((((((.((.((((((....)))))).))))))))))))((((((((......)))))...)))..)))).)))))..)))....... ( -32.70) >DroSec_CAF1 95511 117 + 1 CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUUGGCGCACGCGGUCGAAUUCGCGGAUUGCAGGGGUGGAUAGCAAAGUGGUCUG .........((..((((((((((((((.(((((((.((.((((((....)))))).)))))))))......(((((......))))).....)))))))))).......))))..)) ( -33.51) >DroSim_CAF1 98888 117 + 1 CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUUGGCGCACGCGGUCGAAUUCGCGGAUUGCAGGGGUGGAUAGCAAAAUGGUUUG .............(((((((........(((((((.((.((((((....)))))).))))))))).(((..(((((......)))))...))).)))))))................ ( -32.80) >DroEre_CAF1 102180 117 + 1 CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUUGGGGCACGCGGCCGAAUUCGCGGAUUGCAGGGUUGGAUAACAAAGUGGUUUG (((..(((((((((((((((((((....)))....(((.((((((....)))))).))).....))))))..(((((((......))).)))).))))).)))))..)))....... ( -37.30) >DroYak_CAF1 105720 117 + 1 CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUCGGGGCACGCGGCCGAAUUCGCGAAUGGCUGGGGUGGCUAACAAAGUGGUUUG (((..((((((.((((..((((((.((((((((((.((.((((((....)))))).)))))))))..(((.....)))....))).)))))).)))).))).)))..)))....... ( -34.00) >consensus CUUCUUUAUCAACCCGCUCCAUUCUUGAGAAAUAUUGCUAUGCAAAUAUUUGUAUUGCAUAUUUUGGCGCACGCGGUCGAAUUCGCGGAUUGCAGGGGUGGAUAGCAAAGUGGUUUG .........(((.((((((((((((((.(((((((.((.((((((....)))))).)))))))))......(((((......))))).....))))))))).......))))).))) (-29.83 = -30.67 + 0.84)

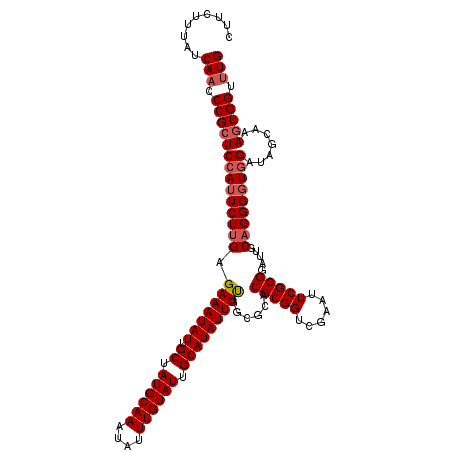
| Location | 16,040,618 – 16,040,735 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -26.84 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
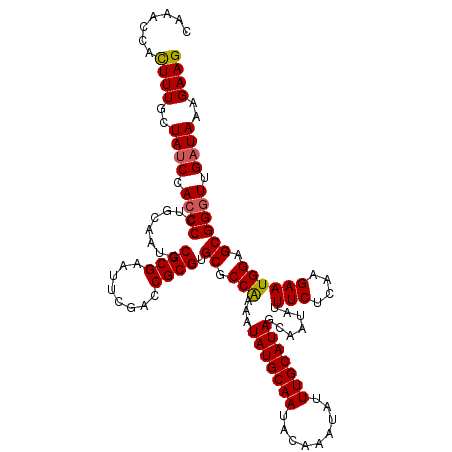
>2R_DroMel_CAF1 16040618 117 - 20766785 CAAACCACUUUGCUAUCUACCCCUGCAAUUCGCGAAUUUGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......((((..((((.(((((((((...((((........))))))).(((...(((((((.........))))))).......(((....)))))))).)))).))))..)))) ( -29.50) >DroSec_CAF1 95511 117 - 1 CAGACCACUUUGCUAUCCACCCCUGCAAUCCGCGAAUUCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......((((..((((.((((........((((........)))).((.(((...(((((((.........))))))).......(((....)))))).)))))).))))..)))) ( -29.70) >DroSim_CAF1 98888 117 - 1 CAAACCAUUUUGCUAUCCACCCCUGCAAUCCGCGAAUUCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......((((..((((.((((........((((........)))).((.(((...(((((((.........))))))).......(((....)))))).)))))).)))).)))). ( -27.80) >DroEre_CAF1 102180 117 - 1 CAAACCACUUUGUUAUCCAACCCUGCAAUCCGCGAAUUCGGCCGCGUGCCCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......((((.(((((.(((((.......((((........)))).((.(((...(((((((.........))))))).......(((....)))))).)))))))))))).)))) ( -30.80) >DroYak_CAF1 105720 117 - 1 CAAACCACUUUGUUAGCCACCCCAGCCAUUCGCGAAUUCGGCCGCGUGCCCCGAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......(((((((((((..(....(((((((((........))).......(((((((((.((.(((....))).)).)).)))))))....)))))).)..)))))))))))... ( -33.30) >consensus CAAACCACUUUGCUAUCCACCCCUGCAAUCCGCGAAUUCGACCGCGUGCGCCAAAAUAUGCAAUACAAAUAUUUGCAUAGCAAUAUUUCUCAAGAAUGGAGCGGGUUGAUAAAGAAG .......((((..((((.((((........((((........)))).((.(((...(((((((.........))))))).......(((....)))))).)))))).))))..)))) (-26.84 = -26.92 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:58 2006