| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,040,401 – 16,040,495 |
| Length | 94 |
| Max. P | 0.843590 |

| Location | 16,040,401 – 16,040,495 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.25 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
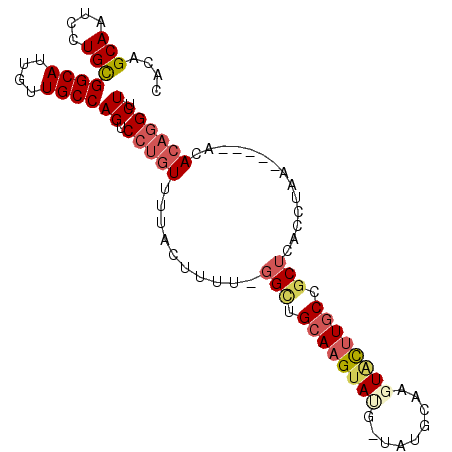
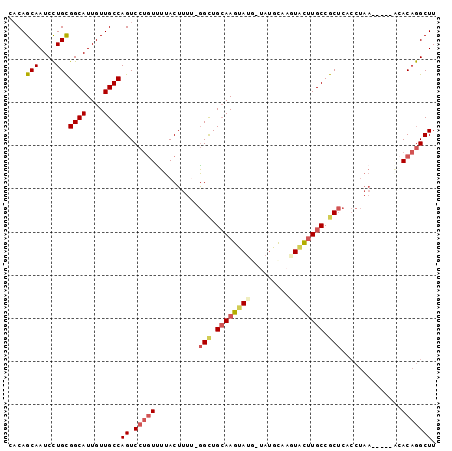
>2R_DroMel_CAF1 16040401 94 + 20766785 AAGCCUGUGU-----UUAGGUGAGCGGCAAGUACUUGCAUAACAUACUUGCAGCCCAAAAGUAAAACAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ..(((((...-----.)))))..(((((((...(((((......(((((.........)))))..........((((....))))))))).))))))). ( -28.40) >DroSec_CAF1 95292 94 + 1 AAGCCUGUGU-----UUAGGUGAGCGGCAAGCACUUGCAUAACAUACUUGCAGCCCAAAAGUAAAACAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ..(((((...-----.)))))..(((((((...(((((......(((((.........)))))..........((((....))))))))).))))))). ( -28.40) >DroSim_CAF1 98668 94 + 1 AAGCCUGUGU-----UUAGGUGAGCGGCAAGCACUUGCAUAACAUACUUGCAGCCCAAAAGUAAAACAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ..(((((...-----.)))))..(((((((...(((((......(((((.........)))))..........((((....))))))))).))))))). ( -28.40) >DroEre_CAF1 101985 88 + 1 AAGCCUGUGC-----UUAGGUGAGCGGCAAGUACUUGCA-----UACUUGCAACC-GAAAGUAAGAGAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ..(((((...-----.)))))..(((((((...(((((.-----..(((((....-....)))))........((((....))))))))).))))))). ( -30.30) >DroYak_CAF1 105505 93 + 1 AAGCCUGUGAGUCUGUUAGGUGAGCGGCAAGUACUUGCA-----UACUUGCAGCC-GAAAGUAAAACAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ...(((((.((((((((......((.(((((((......-----))))))).))(-....)...)))).))))((((....)))))))))......... ( -30.30) >DroAna_CAF1 105925 81 + 1 AAGCUGGUU--------------GUGGCAGAAACUUAUGUA-UGUACUUUCAGCU-AAAAGUAAAACUGGACUGGCAACAAUGCCACAAGAUUGCUG-- .(((...((--------------((((((....((...((.-..((((((.....-.))))))..)).))..((....)).))))))))....))).-- ( -20.70) >consensus AAGCCUGUGU_____UUAGGUGAGCGGCAAGUACUUGCAUA_CAUACUUGCAGCC_AAAAGUAAAACAGGACUGGCAACAAUGCCGCAGGAUUGCUGUG ..(((((.........)))))..(((((((...(((((......(((((.........)))))..........((((....))))))))).))))))). (-22.77 = -23.25 + 0.48)

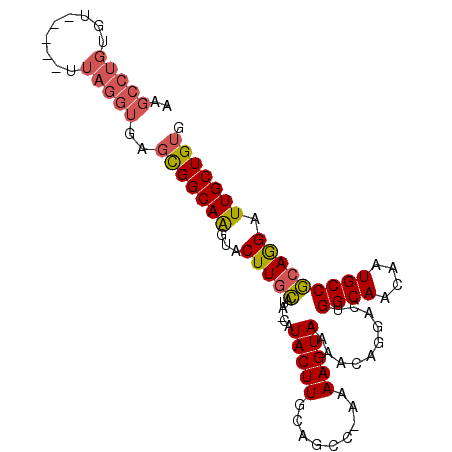
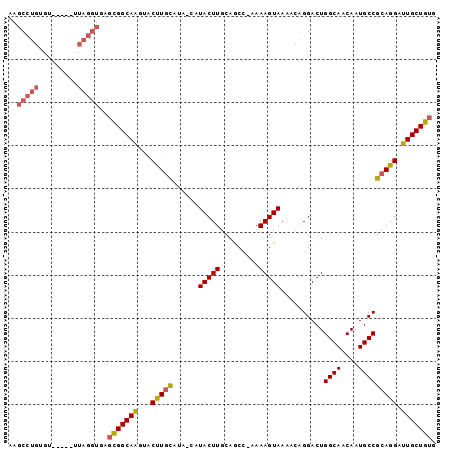
| Location | 16,040,401 – 16,040,495 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.93 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16040401 94 - 20766785 CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUGUUUUACUUUUGGGCUGCAAGUAUGUUAUGCAAGUACUUGCCGCUCACCUAA-----ACACAGGCUU ....(((....)))((((....))))((.(((((((((....(((((.((((((((.........)))))))).)))))..)))-----).))))))). ( -32.20) >DroSec_CAF1 95292 94 - 1 CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUGUUUUACUUUUGGGCUGCAAGUAUGUUAUGCAAGUGCUUGCCGCUCACCUAA-----ACACAGGCUU ....(((....)))((((....))))((.(((((((((....(((((.((((((((.........)))))))).)))))..)))-----).))))))). ( -32.20) >DroSim_CAF1 98668 94 - 1 CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUGUUUUACUUUUGGGCUGCAAGUAUGUUAUGCAAGUGCUUGCCGCUCACCUAA-----ACACAGGCUU ....(((....)))((((....))))((.(((((((((....(((((.((((((((.........)))))))).)))))..)))-----).))))))). ( -32.20) >DroEre_CAF1 101985 88 - 1 CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUCUCUUACUUUC-GGUUGCAAGUA-----UGCAAGUACUUGCCGCUCACCUAA-----GCACAGGCUU ....(((....)))((((....))))((.(((..((((.....-(((.(((((((-----(....)))))))).)))....)))-----)...))))). ( -24.70) >DroYak_CAF1 105505 93 - 1 CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUGUUUUACUUUC-GGCUGCAAGUA-----UGCAAGUACUUGCCGCUCACCUAACAGACUCACAGGCUU .........((((.((((....))))((((.((((........-(((.(((((((-----(....)))))))).))).....))))))))..))))... ( -29.42) >DroAna_CAF1 105925 81 - 1 --CAGCAAUCUUGUGGCAUUGUUGCCAGUCCAGUUUUACUUUU-AGCUGAAAGUACA-UACAUAAGUUUCUGCCAC--------------AACCAGCUU --.(((....(((((((((((....))))...((..(((((((-....)))))))..-.))..........)))))--------------))...))). ( -17.20) >consensus CACAGCAAUCCUGCGGCAUUGUUGCCAGUCCUGUUUUACUUUU_GGCUGCAAGUAUG_UAUGCAAGUACUUGCCGCUCACCUAA_____ACACAGGCUU ....(((....)))((((....))))((.(((((..........(((.((((((((.........)))))))).)))..............))))))). (-18.90 = -19.93 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:55 2006