
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,032,718 – 16,032,815 |
| Length | 97 |
| Max. P | 0.582820 |

| Location | 16,032,718 – 16,032,815 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.02 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16032718 97 + 20766785 CCC---GCCCAGCUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAA-AGGCUCAACUGGCGCAGACUGCGCCUAGACCGAGGCG-------GA-AU------ .((---((((((((...(((((.......)))))....((((((........)))))).-.))))...(((((((.....))))).))...).))))-------).-..------ ( -33.30) >DroPse_CAF1 107815 99 + 1 ---------CUCUUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAAAAGGCUCAAAUGGCGCAGACUGCGCCUAGACUGAGACA-------GAGAUCCCAUA ---------((((....(((((.......)))))....((((((........))))))...(.((((...(((((.....))))).....)))).))-------)))........ ( -24.60) >DroSec_CAF1 87761 92 + 1 --------UCAGCUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAA-AGGCUGAACUGGCGCAGACUCCGCCUAGACCGAGGCG-------GA-AU------ --------((((((...(((((.......)))))....((((((........)))))).-.)))))).(((...)))..(((((((......)))))-------))-..------ ( -29.40) >DroMoj_CAF1 219175 99 + 1 ---------GUCCUGACUUGGCUGUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCA--AGGCUCAACUGGCGCAGACUGCGCCUAGACUAAGGCGC----CCCAGACUCCAA- ---------(((.((....(((((((.(((((((((..............))))))).--((......))))))))).))((((((......))))))----..))))).....- ( -25.74) >DroAna_CAF1 98137 106 + 1 CCCACAACUCUGCUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAA-AGGCUCAACUGGCGCAGACUGCGCCUAGACUAAGGCACAAUCC-GA-AU------ ........(((((....(((((.......)))))....((((((........)))))).-..(((.....)))))))).((.((((......)))).))....-..-..------ ( -23.50) >DroPer_CAF1 101490 99 + 1 ---------CUCUUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAAAAGGCUCAAAUGGCGCAGACUGCGCCUAGACUGAGACA-------GAGAUCCCAUA ---------((((....(((((.......)))))....((((((........))))))...(.((((...(((((.....))))).....)))).))-------)))........ ( -24.60) >consensus _________CUCCUAACAUGGCUAUGACUGCUAUAAAAUGUCAUCUUUUAUUAUGGCAA_AGGCUCAACUGGCGCAGACUGCGCCUAGACUGAGGCA_______GA_AU______ ................(.((((((....((((((((..............))))))))............(((((.....)))))))).))).)..................... (-17.44 = -17.02 + -0.42)

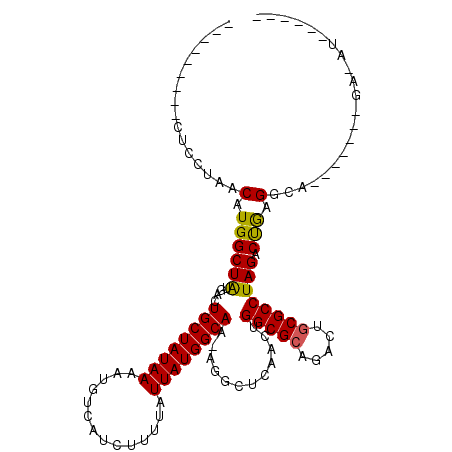
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:52 2006