| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,015,577 – 16,015,682 |
| Length | 105 |
| Max. P | 0.512076 |

| Location | 16,015,577 – 16,015,682 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
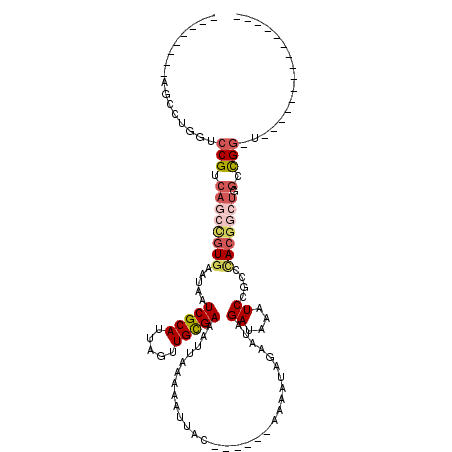
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -11.55 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.95 |
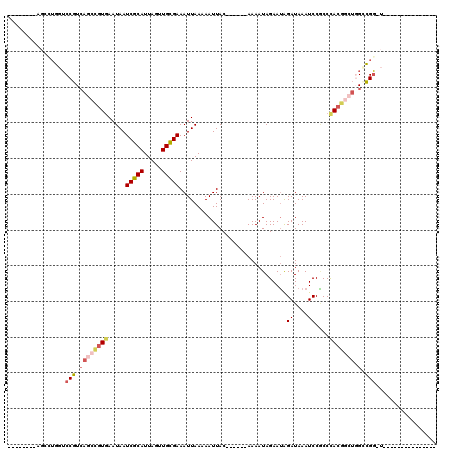
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
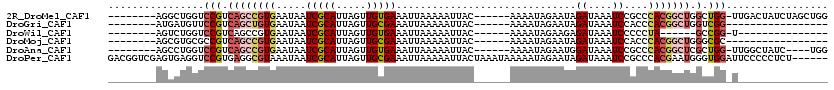
>2R_DroMel_CAF1 16015577 105 - 20766785 --------AGGCUGGUCCGUCAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUGG-UUGACUAUCUAGCUGG --------.((((((.((((((((((((.....(((((.....))))).............------...........((....))....)))))))))).))-........)))))).. ( -30.10) >DroGri_CAF1 174235 89 - 1 --------AUGAUGUUCCGUCAGCUGUGAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCACCCACGGCUGGUCGG----------------- --------........((((((((((((.....(((((.....))))).............------...........((....))....))))))))).)))----------------- ( -22.70) >DroWil_CAF1 264962 84 - 1 --------AGUCUGGUCCGUCAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAGAGAUAAAUCCCCCUA------GCCGG-U--------------- --------...(((((.........((((....)))).....((((((.........))))------)).......................------)))))-.--------------- ( -10.20) >DroMoj_CAF1 191829 89 - 1 --------AGCGUGCGCCGUCAGCCGUGAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCACCCACGGCUGGGCGC----------------- --------.....(((((..((((((((.....(((((.....))))).............------...........((....))....)))))))))))))----------------- ( -28.60) >DroAna_CAF1 79387 101 - 1 --------AGCCUGGUCCGUCAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUGGAUAAAUCCGCCCACGGCUCGCUGG-UUGGCUAUC----UGG --------((((....((((.(((((((.....(((((.....))))).............------..........(((....)))...))))))).)).))-..))))...----... ( -29.00) >DroPer_CAF1 83695 114 - 1 GACGGUCGAGUGAGGUCCGUGAGGCGUAAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUACUAAAUAAAAAUAGAAUAGAUAAAUCCGCCCACGAAUGGGUGGAUUCCCCCUCU------ .((((((......)).))))((((.........(((((.....)))))..................................((((((((((....))))))))))...)))).------ ( -31.70) >consensus ________AGCCUGGUCCGUCAGCCGUGAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUAC______AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCCGG_U_______________ ................(((.((((((((.....(((((.....)))))..............................((....))....))))))).).)))................. (-11.55 = -12.50 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:46 2006