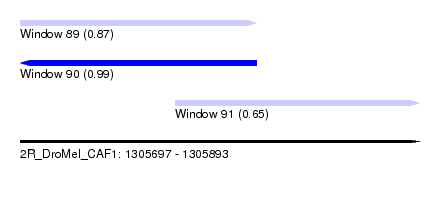
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,305,697 – 1,305,893 |
| Length | 196 |
| Max. P | 0.992099 |

| Location | 1,305,697 – 1,305,813 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -42.75 |
| Consensus MFE | -39.61 |
| Energy contribution | -40.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1305697 116 + 20766785 UAGCUACCAAGGAAGCCAAGCGGCAGGUACGUUGUACACCACACGAACGUCAUUAGCGUCCAGGUGAGGUACACAUACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAA ..(((.((......(((....))).((((.((.((((..(((..(.((((.....)))).)..)))..)))))).)))))).((...((((....)))).)))))........... ( -36.90) >DroSec_CAF1 24078 116 + 1 UAGCUGCCCAGGAAGCCAAGCGGCAGGUACGUUGUGCACCACACGAACGUCAUGAGCGUCCAGGUGAGGUACACAUACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAA ..(((((((......((((((((..((((.((.((((..(((..(.((((.....)))).)..)))..)))))).))))((......)))))))))).)))))))........... ( -45.60) >DroSim_CAF1 3070 116 + 1 UAGCUGCCCAGGAAGCCAAGCGGCAGGUACGUUGUGCACCACACGAACGUCAUGAGCGUCCAGGUGAGGUACACAUACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAA ..(((((((......((((((((..((((.((.((((..(((..(.((((.....)))).)..)))..)))))).))))((......)))))))))).)))))))........... ( -45.60) >DroEre_CAF1 6858 116 + 1 UAGCUGCCAAGGAAGCCGAGCGGCAGAUAUGUGGUACACCACACGAACGUCAUGAGCGUCCAGGUGAGGUACACAUACCGGAGCACGCCCUGCUUGGGGGGCAGCCAUCCAAUGAA ..((((((......(((....)))...((((((.(((..(((..(.((((.....)))).)..)))..)))))))))((.(((((.....))))).)).))))))........... ( -42.90) >consensus UAGCUGCCAAGGAAGCCAAGCGGCAGGUACGUUGUACACCACACGAACGUCAUGAGCGUCCAGGUGAGGUACACAUACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAA ..(((((((......((((((((..((((.((.((((..(((..(.((((.....)))).)..)))..)))))).))))((......)))))))))).)))))))........... (-39.61 = -40.17 + 0.56)

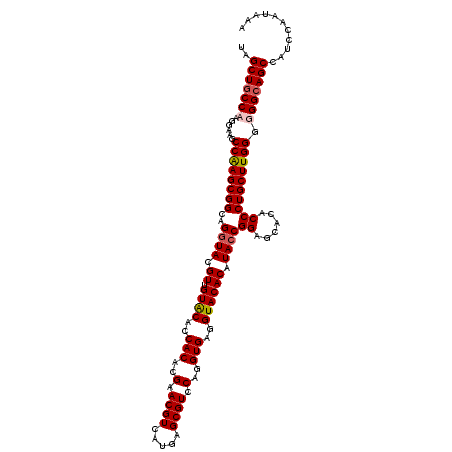
| Location | 1,305,697 – 1,305,813 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -52.70 |
| Consensus MFE | -50.64 |
| Energy contribution | -50.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1305697 116 - 20766785 UUUAUUGGAUGGCUGCCCCCCAAGCAGGGUGUGCUCCGGUAUGUGUACCUCACCUGGACGCUAAUGACGUUCGUGUGGUGUACAACGUACCUGCCGCUUGGCUUCCUUGGUAGCUA .........((((((((....((((..((((.((...(((((((((((.((((.(((((((....).)))))).)))).)))).))))))).))))))..))))....)))))))) ( -49.70) >DroSec_CAF1 24078 116 - 1 UUUAUUGGAUGGCUGCCCCCCAAGCAGGGUGUGCUCCGGUAUGUGUACCUCACCUGGACGCUCAUGACGUUCGUGUGGUGCACAACGUACCUGCCGCUUGGCUUCCUGGGCAGCUA .........(((((((((...((((..((((.((...((((((((((((.(((..((((((....).)))))))).))))))))...)))).))))))..))))...))))))))) ( -54.30) >DroSim_CAF1 3070 116 - 1 UUUAUUGGAUGGCUGCCCCCCAAGCAGGGUGUGCUCCGGUAUGUGUACCUCACCUGGACGCUCAUGACGUUCGUGUGGUGCACAACGUACCUGCCGCUUGGCUUCCUGGGCAGCUA .........(((((((((...((((..((((.((...((((((((((((.(((..((((((....).)))))))).))))))))...)))).))))))..))))...))))))))) ( -54.30) >DroEre_CAF1 6858 116 - 1 UUCAUUGGAUGGCUGCCCCCCAAGCAGGGCGUGCUCCGGUAUGUGUACCUCACCUGGACGCUCAUGACGUUCGUGUGGUGUACCACAUAUCUGCCGCUCGGCUUCCUUGGCAGCUA .........((((((((....((((.(((((.((...(((((((((((.((((.(((((((....).)))))).)))).))).)))))))).))))))).))))....)))))))) ( -52.50) >consensus UUUAUUGGAUGGCUGCCCCCCAAGCAGGGUGUGCUCCGGUAUGUGUACCUCACCUGGACGCUCAUGACGUUCGUGUGGUGCACAACGUACCUGCCGCUUGGCUUCCUGGGCAGCUA .........(((((((((...((((.(((((.((...(((((((((((.((((.(((((((....).)))))).)))).)))).))))))).))))))).))))...))))))))) (-50.64 = -50.70 + 0.06)


| Location | 1,305,773 – 1,305,893 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -48.20 |
| Consensus MFE | -45.15 |
| Energy contribution | -45.90 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1305773 120 + 20766785 ACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAACUUCAUGGCGCGGUAAAGGUAGAUGCAACCAUCUCGGGACCGCUUCUGCUCCGUGAGCGGAGCGGGACGUAUUAGCUCGA .((.(((((.....))))).))((((.(((((............)))))(((((....(.(((((....))))))...)))))((((((((((....)))))))))).......)))).. ( -48.90) >DroSec_CAF1 24154 120 + 1 ACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAACUUCAUGGCGCGGUAAAGGUAGAUGCAACCAUCUCGGGACCUCUUCUGCUCCGUGAGCGGAGCGGGACGUAUCAGCUCAA .((.(((((.....))))).))((((.(((((............)))))(((....(((((((((....)))))....)))).((((((((((....)))))))))))))....)))).. ( -45.70) >DroSim_CAF1 3146 120 + 1 ACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAACUUCAUGGCGCGGUAAAGGUAGAUGCAACCAUCUCGGGACCGCUUCUGCUCCGUGAGCGGAGCGGGACGUAUCAGCUCAA .((.(((((.....))))).))((((.(((((............)))))(((((....(.(((((....))))))...)))))((((((((((....)))))))))).......)))).. ( -48.90) >DroEre_CAF1 6934 120 + 1 ACCGGAGCACGCCCUGCUUGGGGGGCAGCCAUCCAAUGAACUUCAUGGCGCGGUAAAGGUAGAUGCAGCCAUCGCGGGACCGCUUCUGCUCCGUGAGCGGAGCGGGACGUAUCAGCUGAA ((((..((.(.(((.....))).))).(((((............))))).))))...(((.(((((.......(((....)))((((((((((....)))))))))).))))).)))... ( -49.30) >consensus ACCGGAGCACACCCUGCUUGGGGGGCAGCCAUCCAAUAAACUUCAUGGCGCGGUAAAGGUAGAUGCAACCAUCUCGGGACCGCUUCUGCUCCGUGAGCGGAGCGGGACGUAUCAGCUCAA .((.(((((.....))))).))((((.(((((............)))))(((((...(..(((((....))))))...)))))((((((((((....)))))))))).......)))).. (-45.15 = -45.90 + 0.75)

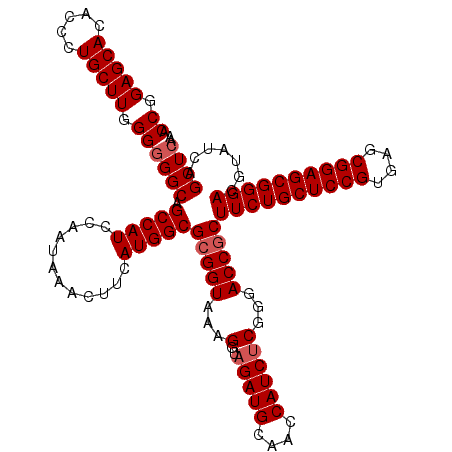
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:45 2006