
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
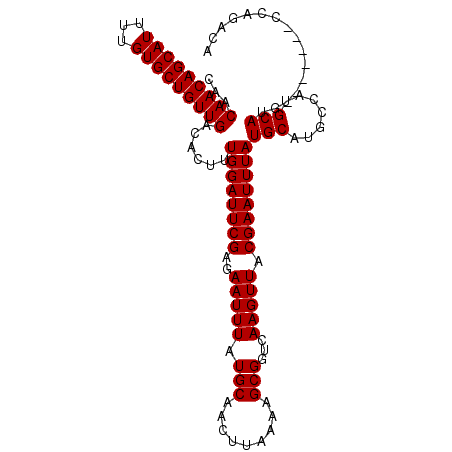
| Location | 16,009,706 – 16,009,813 |
| Length | 107 |
| Max. P | 0.826062 |

| Location | 16,009,706 – 16,009,813 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
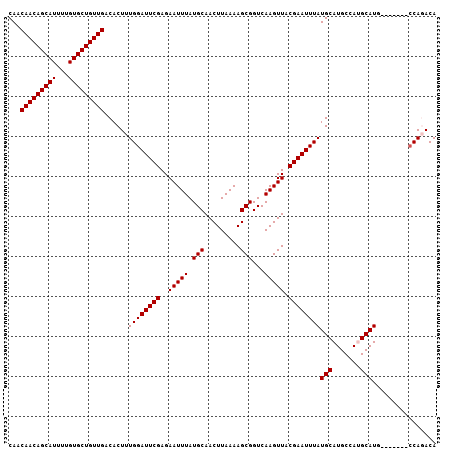
>2R_DroMel_CAF1 16009706 107 + 20766785 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCAUGCAUUCACUCCACCAGACA ...(((((((((...)))))))))....((((((((((..(((((.(((.........)))...))))).)))))..((((((...))))))........))))).. ( -24.60) >DroPse_CAF1 72018 100 + 1 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCACGCAUG-------CCACACA ...(((((((((...)))))))))......((((((((..(((((.(((.........)))...))))).)))))))).((((((...)))))-------)...... ( -27.10) >DroSim_CAF1 65862 99 + 1 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCAUGCAUG--------CAGACA ...(((((((((...)))))))))......((((((((..(((((.(((.........)))...))))).))))))))(((((((...)))))--------)).... ( -29.00) >DroEre_CAF1 71337 107 + 1 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCAUGCAUUCACUCCACCAGACA ...(((((((((...)))))))))....((((((((((..(((((.(((.........)))...))))).)))))..((((((...))))))........))))).. ( -24.60) >DroYak_CAF1 74157 107 + 1 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCAUGCAUUCACUCCACCAGACA ...(((((((((...)))))))))....((((((((((..(((((.(((.........)))...))))).)))))..((((((...))))))........))))).. ( -24.60) >DroPer_CAF1 72493 100 + 1 CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCACGCAUG-------CCACACA ...(((((((((...)))))))))......((((((((..(((((.(((.........)))...))))).)))))))).((((((...)))))-------)...... ( -27.10) >consensus CAACAACAGCAUUUUGUGCUGUUGACACUUUGGAUUCGAGAAUUUAUGCAACUUAAAAGCGGUCAAGUUACGAAUUUAUGCAUGCCAUGCAUG_______CCAGACA ...(((((((((...)))))))))......((((((((..(((((.(((.........)))...))))).))))))))(((.......)))................ (-21.60 = -21.60 + -0.00)

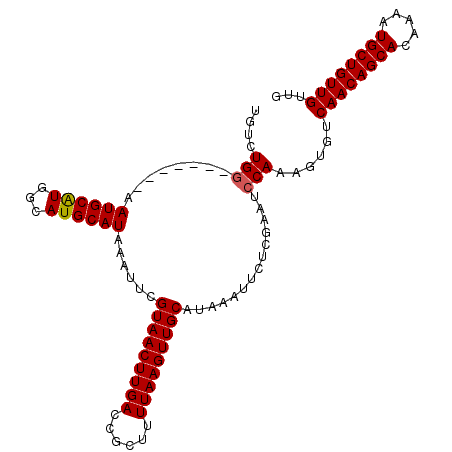
| Location | 16,009,706 – 16,009,813 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.75 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
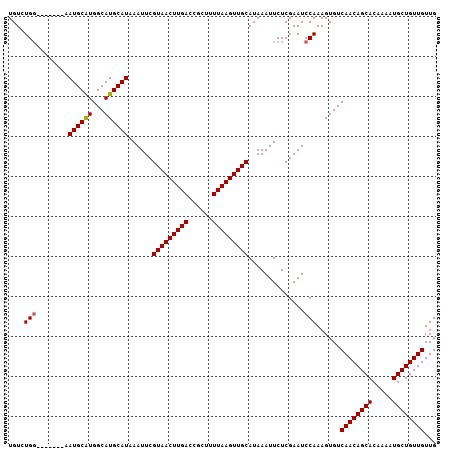
>2R_DroMel_CAF1 16009706 107 - 20766785 UGUCUGGUGGAGUGAAUGCAUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ....(((..((((..((((((...))))))..))))(((((((((......)))))))))..............)))......((((((((.....))))))))... ( -28.70) >DroPse_CAF1 72018 100 - 1 UGUGUGG-------CAUGCGUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ......(-------(((((...))))))........(((((((((......))))))))).......................((((((((.....))))))))... ( -26.90) >DroSim_CAF1 65862 99 - 1 UGUCUG--------CAUGCAUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ....((--------(((((...))))))).......(((((((((......))))))))).......................((((((((.....))))))))... ( -27.50) >DroEre_CAF1 71337 107 - 1 UGUCUGGUGGAGUGAAUGCAUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ....(((..((((..((((((...))))))..))))(((((((((......)))))))))..............)))......((((((((.....))))))))... ( -28.70) >DroYak_CAF1 74157 107 - 1 UGUCUGGUGGAGUGAAUGCAUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ....(((..((((..((((((...))))))..))))(((((((((......)))))))))..............)))......((((((((.....))))))))... ( -28.70) >DroPer_CAF1 72493 100 - 1 UGUGUGG-------CAUGCGUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ......(-------(((((...))))))........(((((((((......))))))))).......................((((((((.....))))))))... ( -26.90) >consensus UGUCUGG_______AAUGCAUGGCAUGCAUAAAUUCGUAACUUGACCGCUUUUAAGUUGCAUAAAUUCUCGAAUCCAAAGUGUCAACAGCACAAAAUGCUGUUGUUG ....(((........((((((...))))))......(((((((((......)))))))))..............)))......((((((((.....))))))))... (-23.81 = -23.75 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:45 2006