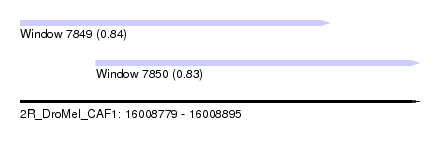
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,008,779 – 16,008,895 |
| Length | 116 |
| Max. P | 0.838229 |

| Location | 16,008,779 – 16,008,869 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -18.77 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
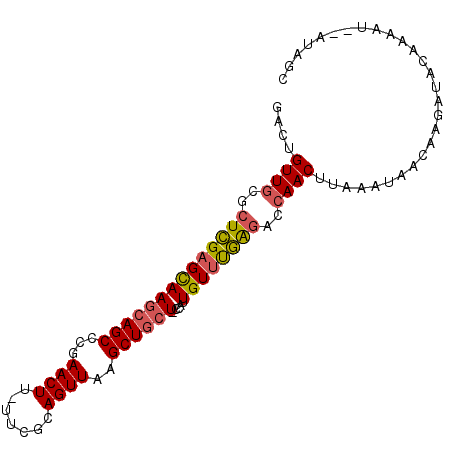
>2R_DroMel_CAF1 16008779 90 + 20766785 -------CGUUGUUUGAG----UGUAUAUUGGCGACUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCUUGUUUUGUUCGAGACCAAC -------.((((((((((----((((....(....)...)))))))(((((((((((...((((.-.....))))..)))))))))))......))).)))) ( -30.80) >DroSec_CAF1 63950 89 + 1 -------CGUUGUUUGAG----UGUAUAUUGGCGACUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCU-GCAUUGUUUGAGACCAAC -------...((((((((----((((....(....)...))))))))))))((((((...((((.-.....))))..))))))-.................. ( -28.90) >DroSim_CAF1 64955 89 + 1 -------CCUUGUUUGAG----UGUAUAUUGGCGAAUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCU-CCAUUGUUUGAGACCAAC -------..(((((((((----((((((((....)))).)))))))))))))(((((...((((.-.....))))..))))).-.................. ( -27.90) >DroEre_CAF1 70410 89 + 1 -------CGUUGUUUGAG----UGUAUAUUGGUGACUGUUGCGCUCGAGCAAGCAGCACAAACUU-UUCGCAGUUAAGCUGCU-CCAUUGUUUGGGACCAAC -------..(((((((((----((((....(....)...)))))))))))))(((((...((((.-.....))))..)))))(-(((.....))))...... ( -31.10) >DroYak_CAF1 73183 96 + 1 CUUCUUUUUUUGUUCGAG----UGUAUAUUGGUGACUGUUGCGCUCGAGCAAGCAGCACAAACUU-UUCGCAGUUAAGCUGCU-CCAUUGUUUGAGACCAAC ..((((...(((((((((----((((....(....)...)))))))))))))(((((...((((.-.....))))..))))).-.........))))..... ( -31.60) >DroAna_CAF1 72794 95 + 1 ------UGUUUGUGCAACUGCUUCUGUAUUGGCGACUGUUGCACUUGAGUAAGCAGCGACAACUUUUUUCCAGUUAAGCUCGU-CUUUUGUCCAAAAUAAAC ------.....(((((((((((........))))...)))))))(((..((((..((((.((((.......))))....))))-..))))..)))....... ( -19.00) >consensus _______CGUUGUUUGAG____UGUAUAUUGGCGACUGUUGCGCUCGAGCAAGCAGCCCGAACUU_UUCGCAGUUAAGCUGCU_CCAUUGUUUGAGACCAAC .....................................((((..((((((((((((((...((((.......))))..)))))).....))))))))..)))) (-18.77 = -18.83 + 0.06)


| Location | 16,008,801 – 16,008,895 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16008801 94 + 20766785 GACUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCUUGUUUUGUUCGAGACCAACUUAAAUAAGAAGACGCAAGAU--AUAGC ...(.(((((.(((((((((((((...((((.-.....))))..)))))))))))((((..((.....))..))))....))))))).).--..... ( -28.50) >DroSec_CAF1 63972 93 + 1 GACUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCU-GCAUUGUUUGAGACCAACUUAAAGAACAAGAUACAAGAU--AUAGC ....((((.(((((((((((((((...((((.-.....)))).....)))-)).))))))))).))))).....................--..... ( -25.80) >DroSim_CAF1 64977 93 + 1 GAAUGUUGCGCUCGAGCAAGCAGCCCGAACUU-UUCGUAGUUAAGCUGCU-CCAUUGUUUGAGACCAACUUAAAUAACAAGAUACAAGAU--AUAGC ....((((.(((((((((((((((...((((.-.....))))..))))))-....)))))))).))))).....................--..... ( -25.10) >DroEre_CAF1 70432 95 + 1 GACUGUUGCGCUCGAGCAAGCAGCACAAACUU-UUCGCAGUUAAGCUGCU-CCAUUGUUUGGGACCAACUUAAACAACAAGAUACAAAAUGCAUAGC ....((((.(((((((((((((((...((((.-.....))))..))))))-....)))))))).)))))............................ ( -22.40) >DroYak_CAF1 73212 93 + 1 GACUGUUGCGCUCGAGCAAGCAGCACAAACUU-UUCGCAGUUAAGCUGCU-CCAUUGUUUGAGACCAACUUGAAUAGCAAGAUACAAAAU--AUAGC ....((((.(((((((((((((((...((((.-.....))))..))))))-....)))))))).))))).....................--..... ( -23.30) >DroAna_CAF1 72821 87 + 1 GACUGUUGCACUUGAGUAAGCAGCGACAACUUUUUUCCAGUUAAGCUCGU-CUUUUGUCCAAAAUAAACUCGAAUUCCGAGGCAUCAA--------- ...((.(((..(((..((((..((((.((((.......))))....))))-..))))..)))......((((.....))))))).)).--------- ( -15.20) >consensus GACUGUUGCGCUCGAGCAAGCAGCCCGAACUU_UUCGCAGUUAAGCUGCU_CCAUUGUUUGAGACCAACUUAAAUAACAAGAUACAAAAU__AUAGC ....((((..((((((((((((((...((((.......))))..)))))).....))))))))..))))............................ (-19.98 = -20.03 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:43 2006