
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,978,429 – 15,978,572 |
| Length | 143 |
| Max. P | 0.940299 |

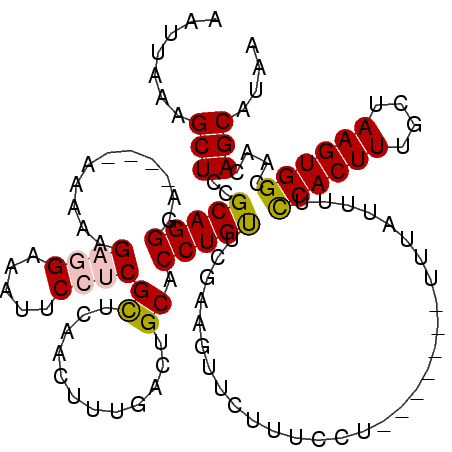
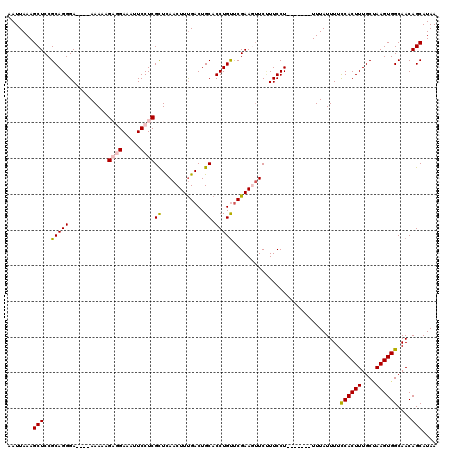
| Location | 15,978,429 – 15,978,538 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15978429 109 + 20766785 AAUUAAAGCUCCGCAGGGA----AAAUAGAGGAAAUUCCUCGUUCAACUUUGACUGCACCUGUUCGAACAUCUUUCCU-------UUUAUUUUCCACUUUGCUAAGUGGCAACAGCAUAA ............(((((((----((...((((.....))))(((((((..((....))...))).))))...))))))-------).......((((((....)))))).....)).... ( -25.50) >DroSec_CAF1 34197 109 + 1 AAUUAAAGCUCCGCAGGGA----AAACAGAGGAAAUUCCUCGCUCAACUUUGACUGCACCUGUUCGAAGUUCUUUCCU-------UUUAUUUUCCACUUUGCUAAGUGGCAACAGCAUAA ............(((((((----((...((((.....))))....((((((((..((....)))))))))).))))))-------).......((((((....)))))).....)).... ( -27.10) >DroSim_CAF1 34370 109 + 1 AAUUAAAGCUCCGCAGGGA----AAACAGAGGAAAUUCCUCGCUCAACUUUGACUGCACCUGUUCGAAGUUCUUUCCU-------UUUAUUUUCCACUUUGCUAAGUGGCAACAGCAUAA ............(((((((----((...((((.....))))....((((((((..((....)))))))))).))))))-------).......((((((....)))))).....)).... ( -27.10) >DroEre_CAF1 40587 109 + 1 AAUUAAAGCUCCGCAGGGA----AAAAAGUUGAAAUUCCUCGCUUAACUUCGACUGCACCUGCCCGAAGUUCGUUCCU-------UUUAUUUUUCACUUUGCAAAGUGGCAACAGCAUAA .......(((..(((((((----(..((((.((......))))))(((((((...((....)).)))))))..)))))-------)........(((((....)))))))...))).... ( -22.10) >DroYak_CAF1 42333 120 + 1 AAUUAAAGCUCCGCAGGGAGAAAAAAAAGUUGAAAUUCCUCGCUUAACUUUGACUGCACCUGUUCGAACUUCCUUCCUCUUAGUUUUUAUUUUUCACUUUGCCAAGUGGCAACAGCAUAA .......(((..(((((..(.....((((((((..........)))))))).....).)))))..(((((...........)))))............(((((....))))).))).... ( -20.40) >consensus AAUUAAAGCUCCGCAGGGA____AAAAAGAGGAAAUUCCUCGCUCAACUUUGACUGCACCUGUUCGAAGUUCUUUCCU_______UUUAUUUUCCACUUUGCUAAGUGGCAACAGCAUAA .......(((..(((((...........((((.....))))((............)).)))))..............................((((((....))))))....))).... (-18.56 = -18.80 + 0.24)

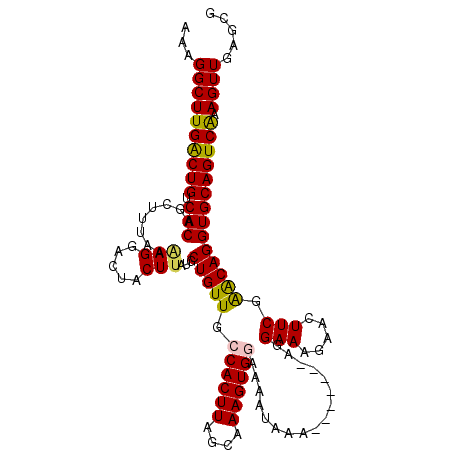
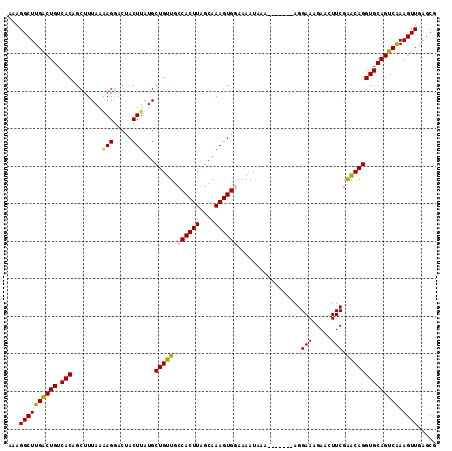
| Location | 15,978,465 – 15,978,572 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15978465 107 - 20766785 AAAGGCUUGACUGUCACAGCUUUAAGAGGACUACUUAUGCUGUUGCCACUUAGCAAAGUGGAAAAUAAA-------AGGAAAGAUGUUCGAACAGGUGCAGUCAAAGUUGAACG ...((((((((((.(((........(((.....)))...(((((.((((((....))))))........-------..(((.....))).))))))))))))).)))))..... ( -26.60) >DroSec_CAF1 34233 107 - 1 AAAGGCUUGACUGUCACAGCUUUAAAAGGACAACUUAUGCUGUUGCCACUUAGCAAAGUGGAAAAUAAA-------AGGAAAGAACUUCGAACAGGUGCAGUCAAAGUUGAGCG ...((((((((((.(((........(((.....)))...(((((.((((((....))))))........-------..(((.....))).))))))))))))).)))))..... ( -26.40) >DroSim_CAF1 34406 107 - 1 AAAGGCUUGACUGUCACAGCUUUAAAAGGACUACUUAUGCUGUUGCCACUUAGCAAAGUGGAAAAUAAA-------AGGAAAGAACUUCGAACAGGUGCAGUCAAAGUUGAGCG ...((((((((((.(((........(((.....)))...(((((.((((((....))))))........-------..(((.....))).))))))))))))).)))))..... ( -26.40) >DroEre_CAF1 40623 106 - 1 GAAGGCUUGGCUGUCACUGCUUUAA-AGUACUACUUAUGCUGUUGCCACUUUGCAAAGUGAAAAAUAAA-------AGGAACGAACUUCGGGCAGGUGCAGUCGAAGUUAAGCG ....((((((((.(((((((....(-((.....)))...(((((..(((((....))))).........-------.....((.....)))))))..))))).)))))))))). ( -27.00) >DroYak_CAF1 42373 114 - 1 AAGGGCUUGACUGUCACUGCUUUAAAAGGACUACUUAUGCUGUUGCCACUUGGCAAAGUGAAAAAUAAAAACUAAGAGGAAGGAAGUUCGAACAGGUGCAGUCAAAGUUAAGCG ....((((((((...(((((.......(((((.(((...((.(((((....)))))(((...........))).))...)))..)))))........)))))...)))))))). ( -32.36) >consensus AAAGGCUUGACUGUCACAGCUUUAAAAGGACUACUUAUGCUGUUGCCACUUAGCAAAGUGGAAAAUAAA_______AGGAAAGAACUUCGAACAGGUGCAGUCAAAGUUGAGCG ...((((((((((.(((........(((.....)))...(((((.((((((....)))))).................(((.....))).)))))))))))))).))))..... (-23.72 = -23.56 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:30 2006