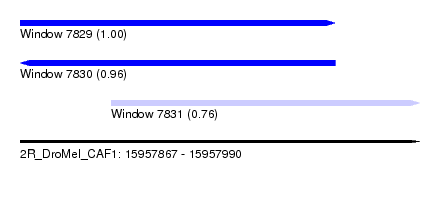
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,957,867 – 15,957,990 |
| Length | 123 |
| Max. P | 0.999584 |

| Location | 15,957,867 – 15,957,964 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -30.36 |
| Energy contribution | -31.64 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15957867 97 + 20766785 AUUGCUGGCAUAAUUAGCCGAGUUGGAGCUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCUAGACUGUGGUCGACUGAAU .....((((.......))))((((((.((.....)).)).(((((..((((((((((...............)))))).)))).))))))))).... ( -31.16) >DroSec_CAF1 13720 91 + 1 AUUGCUGGCAUAAUUAGCCGAGUUGGAGCUGGAGGCG-----CCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACU-CGGUCGGCUGAAU .............((((((((..(.((((((((((((-----((..........)))...(......)....)))))))).))-).))))))))).. ( -35.20) >DroSim_CAF1 13717 97 + 1 AUUGCUGGCAUAAUUAGCCGAGUUGGAGCUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACUGUGGUCGGCUGAAU .............((((((((.(((.((((((((((..(((((.....)))))..((.....))........)))))))).)).))))))))))).. ( -38.40) >DroEre_CAF1 20393 97 + 1 AUUGCUGGCAUAAUUAGCCGAGUUGGAGCAGGAGGCGCCUGGCCAAAAGUCAGAGGAAUACGGCAACAAAAAGCCUCCAGACUGUGGUCGGCUGAAU .............((((((((.(((.(((.((((((..(((((.....)))))........(....).....)))))).).)).))))))))))).. ( -33.40) >DroYak_CAF1 21028 97 + 1 AUUGCUGGCAUAAUUAGCCGAGUUGGAGCUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACGGCAACAAAAAGCCUACAGACUGUGGUCGGCUGAAU .............((((((((.(((.(((((.(((((((((((.....)))..))))....(....).....)))).))).)).))))))))))).. ( -33.80) >consensus AUUGCUGGCAUAAUUAGCCGAGUUGGAGCUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACUGUGGUCGGCUGAAU .............((((((((.(((.((((((((((..(((((.....)))))..((.....))........)))))))).)).))))))))))).. (-30.36 = -31.64 + 1.28)

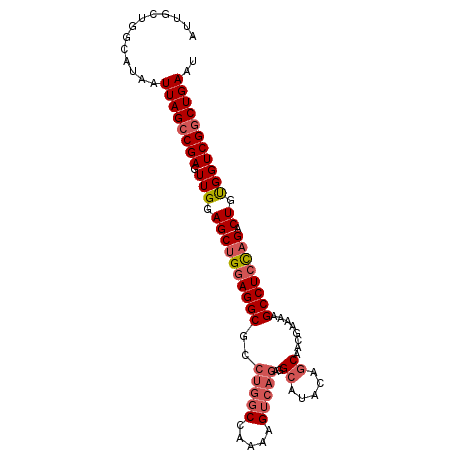
| Location | 15,957,867 – 15,957,964 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15957867 97 - 20766785 AUUCAGUCGACCACAGUCUAGAGGCUUUUCGUUGCUGUAUGCCUCUGACUUUUGGCCAGGCGCCUCCAGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....(((((((((.((((.((((((...............))))))))))..)))...((.((.....)).))...))))))............... ( -26.86) >DroSec_CAF1 13720 91 - 1 AUUCAGCCGACCG-AGUCUGGAGGCUUUUCGUUGCUGUAUGCCUCUGACUUUUGG-----CGCCUCCAGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....((((((..(-((.((((((((.....((......))(((..........))-----))))))))))))....))))))............... ( -31.30) >DroSim_CAF1 13717 97 - 1 AUUCAGCCGACCACAGUCUGGAGGCUUUUCGUUGCUGUAUGCCUCUGACUUUUGGCCAGGCGCCUCCAGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....((((((....((.((((((((........(((....(((..........)))..))))))))))))).....))))))............... ( -30.60) >DroEre_CAF1 20393 97 - 1 AUUCAGCCGACCACAGUCUGGAGGCUUUUUGUUGCCGUAUUCCUCUGACUUUUGGCCAGGCGCCUCCUGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....((((((....(((..((((((..((((..((((...((....))....)))))))).)))))).))).....))))))............... ( -26.50) >DroYak_CAF1 21028 97 - 1 AUUCAGCCGACCACAGUCUGUAGGCUUUUUGUUGCCGUAUGCCUCUGACUUUUGGCCAGGCGCCUCCAGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....((((((....((.(((.((((........(((....(((..........)))..))))))).))))).....))))))............... ( -25.70) >consensus AUUCAGCCGACCACAGUCUGGAGGCUUUUCGUUGCUGUAUGCCUCUGACUUUUGGCCAGGCGCCUCCAGCUCCAACUCGGCUAAUUAUGCCAGCAAU ....(((((((((.((((.((((((....((....))...))))))))))..)))...((.((.....)).))...))))))............... (-24.62 = -24.46 + -0.16)


| Location | 15,957,895 – 15,957,990 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15957895 95 + 20766785 CUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCUAGACUGUGGUCGACUGAAUGGAUUUUCGACCAGAGGUUGCUGUCC ((((((((..(((((.....)))))..((.....))........)))))))).((.(((((((............))))))).)).......... ( -33.40) >DroSec_CAF1 13748 89 + 1 CUGGAGGCG-----CCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACU-CGGUCGGCUGAAUGGAAUUUCGACCAGAGGUUGCUGUCC (((((((((-----((..........)))...(......)....)))))))).((-(((((((............)))))).))).......... ( -28.70) >DroSim_CAF1 13745 95 + 1 CUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACUGUGGUCGGCUGAAUGGAAUUUCGACCAGAGGUUGCUGUCC ((((((((..(((((.....)))))..((.....))........)))))))).((.(((((((............))))))).)).......... ( -34.80) >DroEre_CAF1 20421 95 + 1 CAGGAGGCGCCUGGCCAAAAGUCAGAGGAAUACGGCAACAAAAAGCCUCCAGACUGUGGUCGGCUGAAUGGAAUUUCGACGAGAGGUUGCUGUCC ..(((((((((.(((((..((((.((((.....(....)......))))..)))).)))))))).......((((((.....))))))))).))) ( -31.00) >DroYak_CAF1 21056 95 + 1 CUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACGGCAACAAAAAGCCUACAGACUGUGGUCGGCUGAAUGGAAUUUCGACCAGAGGUUGCUGUCC ..(((((((((((((((..((((..((((....(....).....))))...)))).)))))((.((((......)))).))..))).)))).))) ( -32.00) >consensus CUGGAGGCGCCUGGCCAAAAGUCAGAGGCAUACAGCAACGAAAAGCCUCCAGACUGUGGUCGGCUGAAUGGAAUUUCGACCAGAGGUUGCUGUCC ((((((((..(((((.....)))))..((.....))........)))))))).((.(((((((............))))))).)).......... (-26.12 = -27.60 + 1.48)

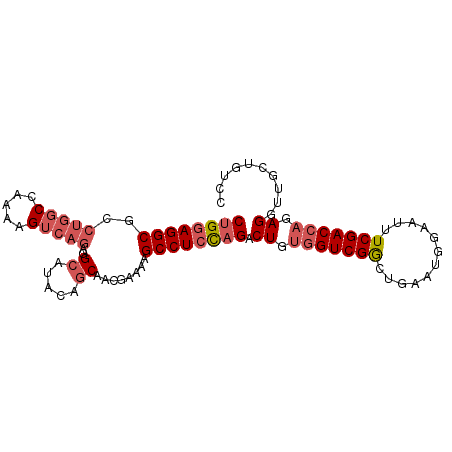
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:24 2006