| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,957,528 – 15,957,628 |
| Length | 100 |
| Max. P | 0.659170 |

| Location | 15,957,528 – 15,957,628 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -20.92 |
| Energy contribution | -22.45 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
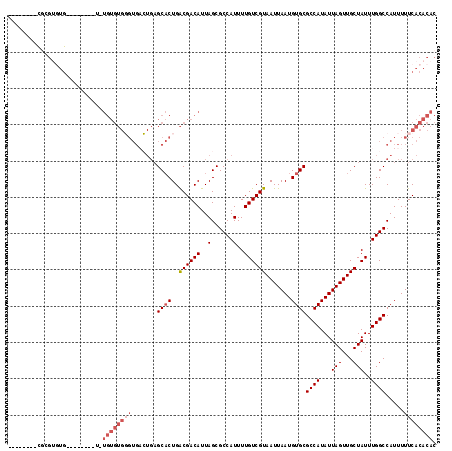
>2R_DroMel_CAF1 15957528 100 + 20766785 --------CGCGUGUG----------UGUGUGGGUGACUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC --------...(((((----------(((((.((((......)))).)))......(.((((....((.((((((((((((...)))))))))))).)).)))))......))))))) ( -31.80) >DroSec_CAF1 13392 102 + 1 --------CGCGUGUG--------UGUGUGUGGGUGACUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC --------...(((((--------(((((((.((((......)))).)).)))...(.((((....((.((((((((((((...)))))))))))).)).)))))......))))))) ( -32.80) >DroSim_CAF1 13388 102 + 1 --------CGCGUAUG--------UCUGUGUGGGUGACUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC --------.((..(((--------((.((((((....))..))))....)))))..))((((....((.((((((((((((...)))))))))))).)).)))).............. ( -28.90) >DroWil_CAF1 160122 112 + 1 UUUCUUUCUCUGUGUGUA------UUUGUGUGCGUGUGUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC ...............(((------(....))))((((((((((((..((((((..(......)..)))))).......))))((((...(((...)))..))))......)))))))) ( -30.60) >DroYak_CAF1 20662 110 + 1 --------CGUGGGUGUAUAUGAGUGUGUGUGGGUGACUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC --------.(((.(((...((((((((.(((.((((......)))).))))))))...((((....((.((((((((((((...)))))))))))).)).)))))))....))).))) ( -32.60) >DroMoj_CAF1 93603 77 + 1 -----------------------------------------GCCCUGGCGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACAUAC -----------------------------------------(((.(((((.(....)))))).......((((((((((((...)))))))))))).....))).............. ( -20.60) >consensus ________CGCGUGUG________U_UGUGUGGGUGACUGAGCACUGACGACAUUAGCGCCAUUUUGUCGUAAUUAAUGUGCGCCAUAUUAGUUGCUAUUUGGCCAUUUUUCACACAC ..........................((((((((.......((((..((((((..(......)..)))))).......))))((((...(((...)))..)))).....)))))))). (-20.92 = -22.45 + 1.53)

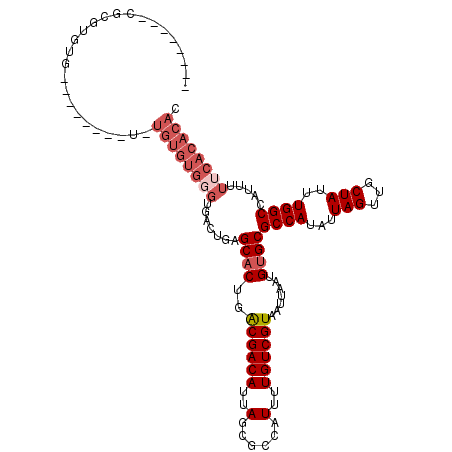
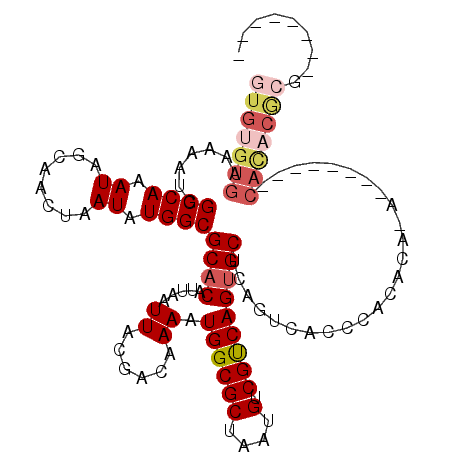
| Location | 15,957,528 – 15,957,628 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -17.80 |
| Energy contribution | -19.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15957528 100 - 20766785 GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCAGUCACCCACACA----------CACACGCG-------- (((((((.....((((.....((((....(((((((((.((((...........))))))))).))))......))))..))))....))))----------))).....-------- ( -26.00) >DroSec_CAF1 13392 102 - 1 GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCAGUCACCCACACACA--------CACACGCG-------- (((((((.....((((.....((((....(((((((((.((((...........))))))))).))))......))))..))))....))))))--------).......-------- ( -26.00) >DroSim_CAF1 13388 102 - 1 GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCAGUCACCCACACAGA--------CAUACGCG-------- (((((((.....(((......((((....(((((((((.((((...........))))))))).))))......)))).......)))......--------))))))).-------- ( -23.22) >DroWil_CAF1 160122 112 - 1 GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCACACACGCACACAAA------UACACACAGAGAAAGAAA (((((((.....((((....(((...)))...((((((.((((...........)))))))))).....))))((((........)))).....------)))))))........... ( -26.80) >DroYak_CAF1 20662 110 - 1 GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCAGUCACCCACACACACUCAUAUACACCCACG-------- (((((((.....((((.....((((....(((((((((.((((...........))))))))).))))......))))..))))....)))))))...............-------- ( -25.50) >DroMoj_CAF1 93603 77 - 1 GUAUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGCCAGGGC----------------------------------------- ((.(((((..((((((((.((........)).))))...))))..)))))))....((((((....).)))))....----------------------------------------- ( -16.30) >consensus GUGUGUGAAAAAUGGCCAAAUAGCAACUAAUAUGGCGCACAUUAAUUACGACAAAAUGGCGCUAAUGUCGUCAGUGCUCAGUCACCCACACA_A________CACACGCG________ (((((((.......((((.((........)).))))((((.....((......)).((((((....).))))))))).........................)))))))......... (-17.80 = -19.05 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:22 2006