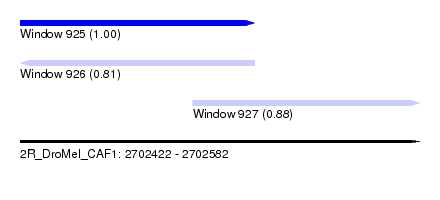
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,702,422 – 2,702,582 |
| Length | 160 |
| Max. P | 0.996540 |

| Location | 2,702,422 – 2,702,516 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2702422 94 + 20766785 AUCGCAGUUAAGAGCAAUGCAAUACAGCAAUGGCAACUUCUGCAUUUGCCUAGGCUGAGGCGUGUGCAAACGCGCAUACGCCCCGUUGGCCUAG ...((((......((..(((......)))...)).....))))......((((((..((((((((((......)))))))))...)..)))))) ( -37.90) >DroSec_CAF1 17329 75 + 1 AUCGCAGUUAGCA-------------------ACAACUUCUGCAUUUGCCUCGGCUGAGGCGUGUGCAAACGCGCAUACGCCCCGUUGGCCUAG ...((((..((..-------------------....)).))))....(((.(((....(((((((((......))))))))))))..))).... ( -29.10) >DroSim_CAF1 14734 75 + 1 AUCGCAGUUAGCA-------------------GCAACUUCUGCAUUUGUCUCGGCUGAGGCGUGUGCAAACGCGCAUACGCCCCGUUGGCCUAG ...((((...(((-------------------(......))))..))))...(((..((((((((((......)))))))))...)..)))... ( -28.80) >DroEre_CAF1 14767 80 + 1 AUCGCAGUCAACAGCACUGC--------AAUGGCAACUUCUGUAUUUU------CCGAGGCGUGUGCGAACGCGCAUACGCCCCAUUGGCCUAG ...((((((....).)))))--------...(((.((....)).....------..(.((((((((((....)))))))))).)....)))... ( -29.40) >DroYak_CAF1 14439 86 + 1 AUUGCAGUUAACAGCAAUUC--------AAUGCCAACUUCUGCAGUUGCUUCGGCUGAGGUGUGUGCAAACGCGCAUACGCCCCGUUGGCCUAG (((((........)))))..--------...((((((..........((....)).(.(((((((((......))))))))).))))))).... ( -31.20) >consensus AUCGCAGUUAACAGCA_U_C________AAUGGCAACUUCUGCAUUUGCCUCGGCUGAGGCGUGUGCAAACGCGCAUACGCCCCGUUGGCCUAG ...((((................................)))).........(((((((((((((((......)))))))))...))))))... (-24.33 = -24.25 + -0.08)

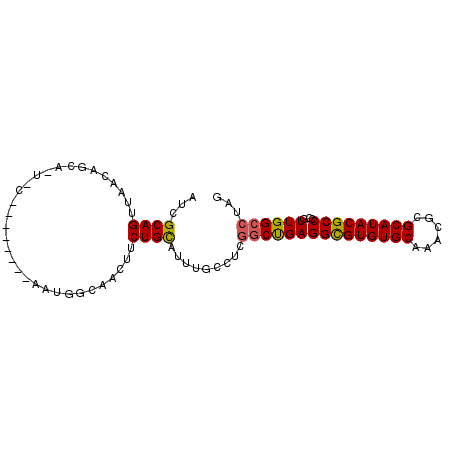
| Location | 2,702,422 – 2,702,516 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2702422 94 - 20766785 CUAGGCCAACGGGGCGUAUGCGCGUUUGCACACGCCUCAGCCUAGGCAAAUGCAGAAGUUGCCAUUGCUGUAUUGCAUUGCUCUUAACUGCGAU ((((((....(((((((.((((....)))).))))))).))))))((((.(((((.(((....))).)))))))))(((((........))))) ( -36.80) >DroSec_CAF1 17329 75 - 1 CUAGGCCAACGGGGCGUAUGCGCGUUUGCACACGCCUCAGCCGAGGCAAAUGCAGAAGUUGU-------------------UGCUAACUGCGAU ((.(((....(((((((.((((....)))).))))))).))).)).....(((((.(((...-------------------.)))..))))).. ( -26.60) >DroSim_CAF1 14734 75 - 1 CUAGGCCAACGGGGCGUAUGCGCGUUUGCACACGCCUCAGCCGAGACAAAUGCAGAAGUUGC-------------------UGCUAACUGCGAU ((.(((....(((((((.((((....)))).))))))).))).)).....(((((.(((...-------------------.)))..))))).. ( -26.40) >DroEre_CAF1 14767 80 - 1 CUAGGCCAAUGGGGCGUAUGCGCGUUCGCACACGCCUCGG------AAAAUACAGAAGUUGCCAUU--------GCAGUGCUGUUGACUGCGAU ...(((.((((((((((.((((....)))).)))))))(.------......)....))))))(((--------(((((.(....))))))))) ( -29.70) >DroYak_CAF1 14439 86 - 1 CUAGGCCAACGGGGCGUAUGCGCGUUUGCACACACCUCAGCCGAAGCAACUGCAGAAGUUGGCAUU--------GAAUUGCUGUUAACUGCAAU ....((((((((((.((.((((....)))).)).)))).......((....))....))))))...--------..(((((........))))) ( -27.40) >consensus CUAGGCCAACGGGGCGUAUGCGCGUUUGCACACGCCUCAGCCGAGGCAAAUGCAGAAGUUGCCAUU________G_A_UGCUGUUAACUGCGAU ...(((....(((((((.((((....)))).))))))).)))........(((((................................))))).. (-18.79 = -19.27 + 0.48)

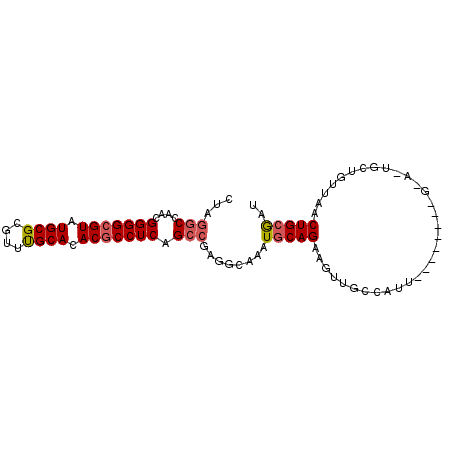
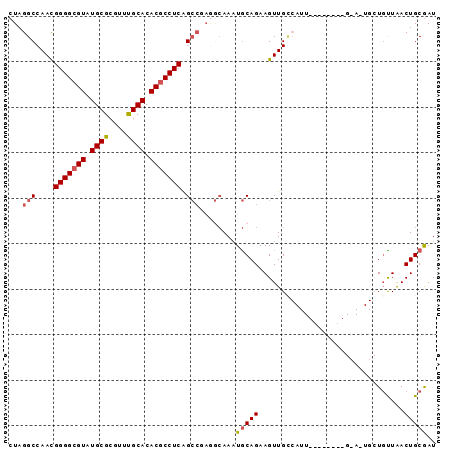
| Location | 2,702,491 – 2,702,582 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2702491 91 + 20766785 ACGCGCAUACGCCCCGUUGGCCUAGUGCGAUUUCAAUCAGCCGCUGCCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCA ..((.((...(((((((((((..((((((((....))).).))))))))))))....(((.(((........))).))))))....)))). ( -29.20) >DroSec_CAF1 17379 91 + 1 ACGCGCAUACGCCCCGUUGGCCUAGUGCGAUUUCAAUCAGCCACUUUCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCA .(((((.((.(((.....))).)))))))..........((((((......))(((.(((.(((........))).))))))....)))). ( -24.90) >DroSim_CAF1 14784 91 + 1 ACGCGCAUACGCCCCGUUGGCCUAGUGCGAUUUCAAUCAGCCACUUCCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCA ..((.((...((((((((((...((((((((....))).).)))).)))))))....(((.(((........))).))))))....)))). ( -26.80) >DroEre_CAF1 14822 91 + 1 ACGCGCAUACGCCCCAUUGGCCUAGUGCGAUUUCAAUCAGCCGCUGCCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCU ..((.((...(((((((((((..((((((((....))).).))))))))))))....(((.(((........))).))))))....)))). ( -29.10) >DroYak_CAF1 14500 91 + 1 ACGCGCAUACGCCCCGUUGGCCUAGUGCGAUUGCAAGCAGCCGCUGCCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCA ..((.((...(((((((((((.....(((..((....))..))).))))))))....(((.(((........))).))))))....)))). ( -31.10) >consensus ACGCGCAUACGCCCCGUUGGCCUAGUGCGAUUUCAAUCAGCCGCUGCCAAUGGGUCAGGCAUUAUCAGAUAUUAACGCCGGCAAUUUGGCA ..((.((...(((((((((((..((((((((....))).).))))))))))))....(((.(((........))).))))))....)))). (-26.68 = -26.72 + 0.04)

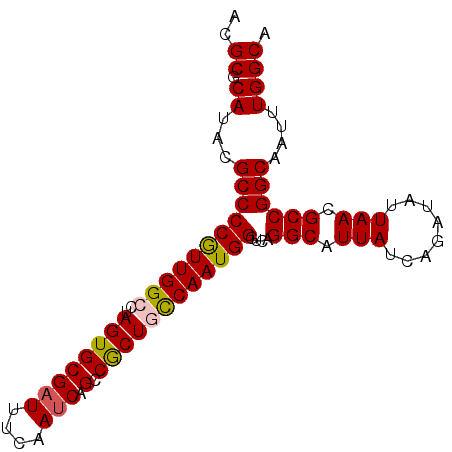
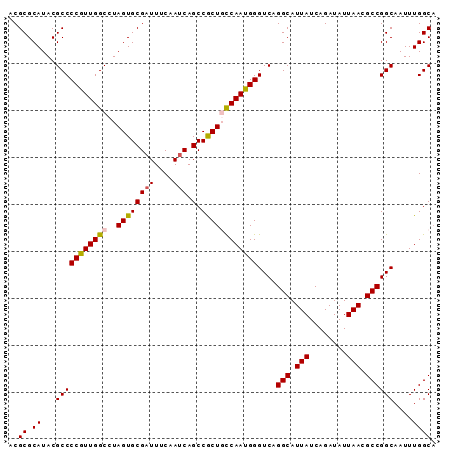
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:55 2006