
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,940,239 – 15,940,346 |
| Length | 107 |
| Max. P | 0.525752 |

| Location | 15,940,239 – 15,940,346 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15940239 107 - 20766785 AAUUUC-----GCCACCAUCAGAGAUCUCAACAGUUUGCUUUGAAGUGGC--AUUCAUC------UGUGUAAUUUCAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAACGUAU .(((((-----..........))))).......(((((...(((((((((--.......------.(((((((...))))))))).)))))))((......))........))))).... ( -20.80) >DroPse_CAF1 72505 109 - 1 ---UUC-----UCCAGCAGAAGAGGCCA-GGCAGCCU--UUUCGAGUGGCACAUUCGACAGACAGUGUGUAAUGAAAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAAUGUAU ---...-----....((.((((((((..-....))))--))))(((((......((....))..(((((((((...))))))))).)))))..))......................... ( -27.50) >DroSec_CAF1 67614 107 - 1 AAUUUC-----GCCACCAUCACAGAUCUCAACAGUUUGCUUUGAAGUGGC--AUUCAUC------UGUGUAAUUUUAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAACGUAU ......-----......................(((((...(((((((((--.......------.(((((((...))))))))).)))))))((......))........))))).... ( -19.80) >DroSim_CAF1 66940 107 - 1 AAUUUC-----GCCACCAUCACAGAUCUCAACAGUUUGCUUUGAAGUGGC--AUUCAUC------UGUGUAAUUUCAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAACGCAU ......-----((..........(....)....(((((...(((((((((--.......------.(((((((...))))))))).)))))))((......))........))))))).. ( -20.10) >DroAna_CAF1 62444 106 - 1 AAUUUGUGUGGGGUAUCAUCAAAGGCCCCAGCACUU-GCA--CAAGUGGC--AUUCAAC------CGUGUAAUGAAAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAAUA--- .((((((.((((((..........))))))((..((-((.--.(((((((--.......------.(((((((...))))))))).)))))..))))....)).......)))))).--- ( -27.20) >DroPer_CAF1 72704 109 - 1 ---UUC-----UCCAGCAGAAGAGGCCA-GGCAGCCU--UUUCGAGUGGCACAUUCGACAGACAGUGUGUAAUGAAAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAAUGUAU ---...-----....((.((((((((..-....))))--))))(((((......((....))..(((((((((...))))))))).)))))..))......................... ( -27.50) >consensus AAUUUC_____GCCACCAUCAGAGACCUCAACAGUUUGCUUUCAAGUGGC__AUUCAAC______UGUGUAAUGAAAUUACACGCACACUUCAGCAAUUUAGCAAUUUAAACAAACGUAU ...........................................(((((((................(((((((...))))))))).)))))..((......))................. (-10.10 = -9.87 + -0.22)

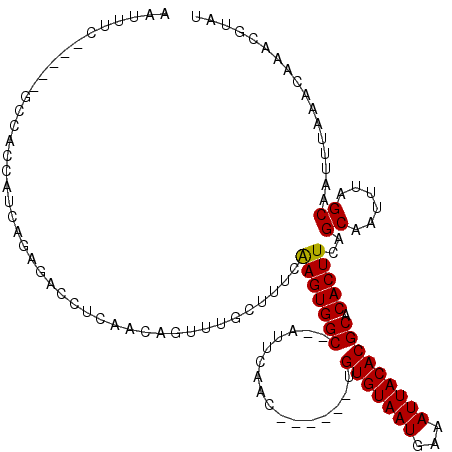
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:13 2006