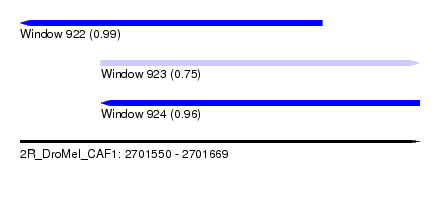
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,701,550 – 2,701,669 |
| Length | 119 |
| Max. P | 0.989359 |

| Location | 2,701,550 – 2,701,640 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2701550 90 - 20766785 AAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGUCCGCCAGCCAAACUAGUUUGGGCGAAAGCCCGGCGAUCUAAUGAGCC ..(((((((((......))))))..............(((((((.(((....(((....)))((((....))))))))))))))..))). ( -33.80) >DroSec_CAF1 16475 90 - 1 AAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGCGAAAGCCAAGAGAUCUAAUGAGCC ..(((..((((((.(..((((...................)))).)))))))....(((.((.(((....)))....)).)))...))). ( -29.71) >DroSim_CAF1 13875 90 - 1 AAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGCGAAAGCCAAGAGAUCUAAUGAGCC ..(((..((((((.(..((((...................)))).)))))))....(((.((.(((....)))....)).)))...))). ( -29.71) >DroEre_CAF1 13890 90 - 1 AAGUUGCGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCGAAACUAGUUUGGGCCAAAGCCAAGCGAUCUAAUGAGCC ...(((((((.(((((((((.((.........)).)))).)))))))).)))).....((((((.......))))))............. ( -28.20) >DroYak_CAF1 13561 90 - 1 AAGUUACGGCUGGGCCAUGCCGUAAUAAUUACAAAGUUUAGGCCUGCCAGCCAAACUAGUUUGGGCGAAAGCCAAGCGAUCUAAUGAGCC ..((((((((........))))))))........(((((.(((......)))))))).((((((.(....)))))))............. ( -29.50) >consensus AAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGCGAAAGCCAAGCGAUCUAAUGAGCC ..(((..(((.(((((((((.(((.....)))...)))).))))))))...............(((....))).............))). (-25.68 = -25.32 + -0.36)

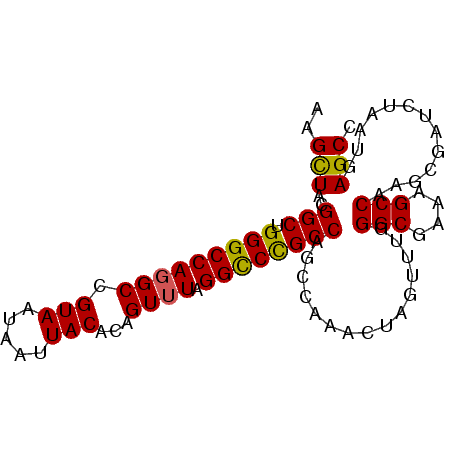
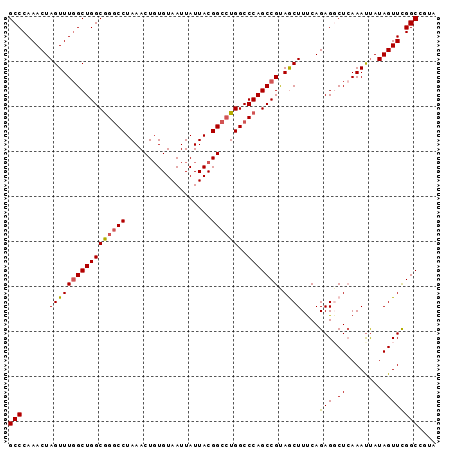
| Location | 2,701,574 – 2,701,669 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -23.93 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2701574 95 + 20766785 GCCCAAACUAGUUUGGCUGGCGGACCUAAACUGUGUAAUUAUUACGGCCUGGCCCAGCCGUAGCUUUCAGAGGCUCAAAUUAUAGUUUGGCAGUA (((.((((((....((((((..(.((....(((((.......)))))...)))))))))(.(((((....))))))......))))))))).... ( -29.70) >DroSec_CAF1 16499 95 + 1 GCCCAAACUAGUUUGGCUGGCGGGCCUAAACUGUGUAAUUAUUACGGCCUGGCCCAGCCGUAGCUUUCAGAGUCUCAAAUUAUAGUUCGGCGGUA (((..(((((((((((..((((((((....(((((.......)))))...))))).)))...((((...)))).))))))..))))).))).... ( -30.20) >DroSim_CAF1 13899 95 + 1 GCCCAAACUAGUUUGGCUGGCGGGCCUAAACUGUGUAAUUAUUACGGCCUGGCCCAGCCGUAGCUUUCAGAGUCUCAAAUUAUAGUUCGGCGGUA (((..(((((((((((..((((((((....(((((.......)))))...))))).)))...((((...)))).))))))..))))).))).... ( -30.20) >DroEre_CAF1 13914 95 + 1 GCCCAAACUAGUUUCGCUGGCGGGCCUAAACUGUGUAAUUAUUACGGCCUGGCCCAGCCGCAACUUUCAGAGGCUCAAACCAUAGUUCGGCCAUA (((..(((((.....((.((((((((....(((((.......)))))...))))).)))))..........((......)).))))).))).... ( -30.40) >DroYak_CAF1 13585 95 + 1 GCCCAAACUAGUUUGGCUGGCAGGCCUAAACUUUGUAAUUAUUACGGCAUGGCCCAGCCGUAACUUCCAGAGGCUAAAAUUAUAGUUCGGCCGUA (((..(((((.((((((.((....))....(((((......(((((((........)))))))....)))))))))))....))))).))).... ( -27.80) >consensus GCCCAAACUAGUUUGGCUGGCGGGCCUAAACUGUGUAAUUAUUACGGCCUGGCCCAGCCGUAGCUUUCAGAGGCUCAAAUUAUAGUUCGGCCGUA (((......(((((((((((((((((...................))))))..))))))).))))....(((.((........)))))))).... (-23.93 = -23.97 + 0.04)

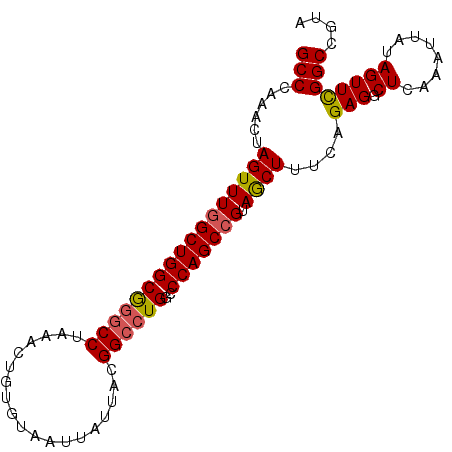
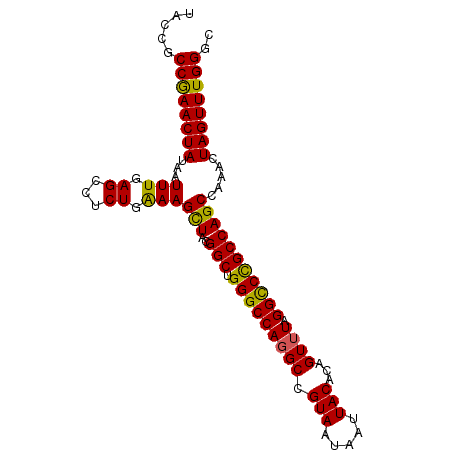
| Location | 2,701,574 – 2,701,669 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -26.40 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2701574 95 - 20766785 UACUGCCAAACUAUAAUUUGAGCCUCUGAAAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGUCCGCCAGCCAAACUAGUUUGGGC .....((((((((...((((.((.......(((....)))(((((((((.((.........)).)))).)))))....)))))).)))))))).. ( -28.00) >DroSec_CAF1 16499 95 - 1 UACCGCCGAACUAUAAUUUGAGACUCUGAAAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGC .....((((((((...((((.(...(((..(((....)))(((((((((.((.........)).)))).)))))..)))))))).)))))))).. ( -28.40) >DroSim_CAF1 13899 95 - 1 UACCGCCGAACUAUAAUUUGAGACUCUGAAAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGC .....((((((((...((((.(...(((..(((....)))(((((((((.((.........)).)))).)))))..)))))))).)))))))).. ( -28.40) >DroEre_CAF1 13914 95 - 1 UAUGGCCGAACUAUGGUUUGAGCCUCUGAAAGUUGCGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCGAAACUAGUUUGGGC ...(.((((((((.(((....)))........(((((((.(((((((((.((.........)).)))).)))))))).))))...)))))))).) ( -33.60) >DroYak_CAF1 13585 95 - 1 UACGGCCGAACUAUAAUUUUAGCCUCUGGAAGUUACGGCUGGGCCAUGCCGUAAUAAUUACAAAGUUUAGGCCUGCCAGCCAAACUAGUUUGGGC ...(.(((((((....((((((...))))))((((((((........))))))))........(((((.(((......))))))))))))))).) ( -28.70) >consensus UACCGCCGAACUAUAAUUUGAGCCUCUGAAAGCUACGGCUGGGCCAGGCCGUAAUAAUUACACAGUUUAGGCCCGCCAGCCAAACUAGUUUGGGC .....((((((((...(((.((...)).)))(((..(((.(((((((((.(((.....)))...)))).))))))))))).....)))))))).. (-26.40 = -25.72 + -0.68)

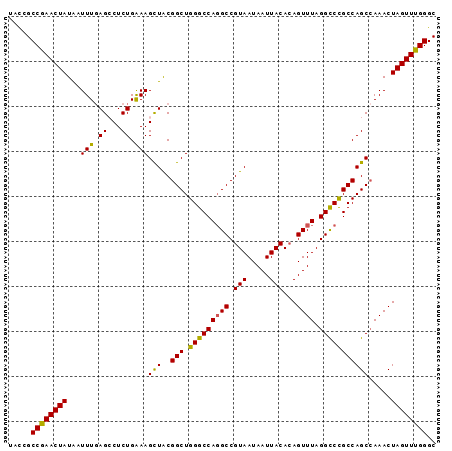
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:52 2006