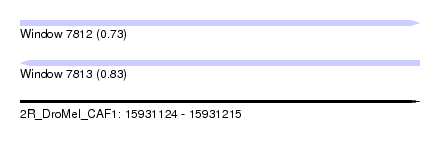
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,931,124 – 15,931,215 |
| Length | 91 |
| Max. P | 0.827145 |

| Location | 15,931,124 – 15,931,215 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15931124 91 + 20766785 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAGCAAAUAAAA-------------------------AGACGGA-AACGUCUAUAUUUUAUUUGCACAACCUGCGGG---CGGUAUUC .(.((((((((..((((.......))))(((((.((((((((((-------------------------(((((..-..)))))...)))))))))).)))))))))))---)).).... ( -33.00) >DroSec_CAF1 58596 91 + 1 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAGCAAAUAGAA-------------------------AGACGGA-AAUGUCUAUAUUUUAUUUGCACAACCUGCGGG---CGGCAUUU ..(((((((((..((((.......))))(((((.((((((((((-------------------------(((((..-..)))))...)))))))))).)))))))))))---).)).... ( -27.20) >DroSim_CAF1 57479 91 + 1 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAGCAAAUAGAA-------------------------AGACGGA-AAUGUCUAUAUUUUAUUUGCACAACCUGCGGG---CGGCAUUU ..(((((((((..((((.......))))(((((.((((((((((-------------------------(((((..-..)))))...)))))))))).)))))))))))---).)).... ( -27.20) >DroEre_CAF1 58264 91 + 1 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAGCAAAUAGAA-------------------------AGACGGA-AACGGCUAUAUUUUAUUUGCACAACCUGGGGG---CGGCAUUU (((((((.((...((((.......))))(((((.((((((((((-------------------------((.((..-..)).))...)))))))))).))))).....)---)))))))) ( -26.20) >DroYak_CAF1 60670 95 + 1 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGUGCAAAUAGAA-------------------------AGACGGAAAACGGCUAUAUUUUAUUUGCACAACCUGCGGGCUGCGGCAUUU (((((((.((((...((((.........((((((((((((((((-------------------------((.((.....)).))...))))))))))))))))))))..))))))))))) ( -31.90) >DroAna_CAF1 53051 117 + 1 GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAACAAAUAGAUGUGCUGCGGGACCCGGGCUAUAUUUAAACAGAUAUUUUCUUUCUUUUAUUUGCCCAACCUGCCGG---CGAAAUAC ..((((((((((.((((.......))))(((((..((((((((((((((.(((...)))))).)))))))...(((.....)))........))))..)))))..)).)---).)))))) ( -22.80) >consensus GAGUGUUUGCGGAAAUGUGAUGAUUAUUGGUUGAGCAAAUAGAA_________________________AGACGGA_AACGUCUAUAUUUUAUUUGCACAACCUGCGGG___CGGCAUUU (((((((..((....))..)........(((((.((((((((((.........................(((((.....)))))...)))))))))).)))))...........)))))) (-17.59 = -17.92 + 0.34)

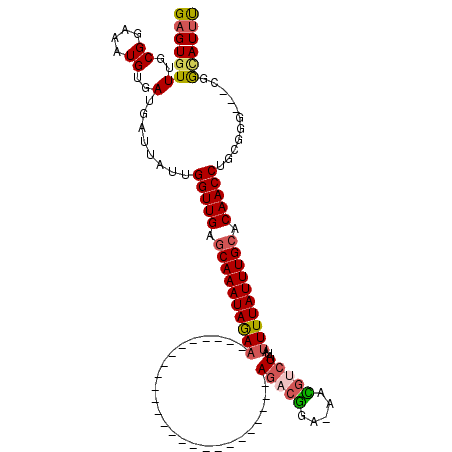
| Location | 15,931,124 – 15,931,215 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -12.30 |
| Energy contribution | -12.16 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15931124 91 - 20766785 GAAUACCG---CCCGCAGGUUGUGCAAAUAAAAUAUAGACGUU-UCCGUCU-------------------------UUUUAUUUGCUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC ........---......(((((.((((((((((...(((((..-..)))))-------------------------)))))))))).)))))............................ ( -23.40) >DroSec_CAF1 58596 91 - 1 AAAUGCCG---CCCGCAGGUUGUGCAAAUAAAAUAUAGACAUU-UCCGUCU-------------------------UUCUAUUUGCUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC ...(((..---...)))(((((.(((((((......((((...-...))))-------------------------...))))))).)))))............................ ( -20.00) >DroSim_CAF1 57479 91 - 1 AAAUGCCG---CCCGCAGGUUGUGCAAAUAAAAUAUAGACAUU-UCCGUCU-------------------------UUCUAUUUGCUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC ...(((..---...)))(((((.(((((((......((((...-...))))-------------------------...))))))).)))))............................ ( -20.00) >DroEre_CAF1 58264 91 - 1 AAAUGCCG---CCCCCAGGUUGUGCAAAUAAAAUAUAGCCGUU-UCCGUCU-------------------------UUCUAUUUGCUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC ........---......(((((.(((((((......((.((..-..)).))-------------------------...))))))).)))))............................ ( -15.30) >DroYak_CAF1 60670 95 - 1 AAAUGCCGCAGCCCGCAGGUUGUGCAAAUAAAAUAUAGCCGUUUUCCGUCU-------------------------UUCUAUUUGCACAACCAAUAAUCAUCACAUUUCCGCAAACACUC ...(((.((.....)).(((((((((((((......((.((.....)).))-------------------------...)))))))))))))..................)))....... ( -23.20) >DroAna_CAF1 53051 117 - 1 GUAUUUCG---CCGGCAGGUUGGGCAAAUAAAAGAAAGAAAAUAUCUGUUUAAAUAUAGCCCGGGUCCCGCAGCACAUCUAUUUGUUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC ........---......(((((((((((((..(((.(((.....))).))).......((.(((...)))..)).....)))))))))))))............................ ( -20.00) >consensus AAAUGCCG___CCCGCAGGUUGUGCAAAUAAAAUAUAGACAUU_UCCGUCU_________________________UUCUAUUUGCUCAACCAAUAAUCAUCACAUUUCCGCAAACACUC .................(((((.(((((((.................................................))))))).)))))............................ (-12.30 = -12.16 + -0.14)
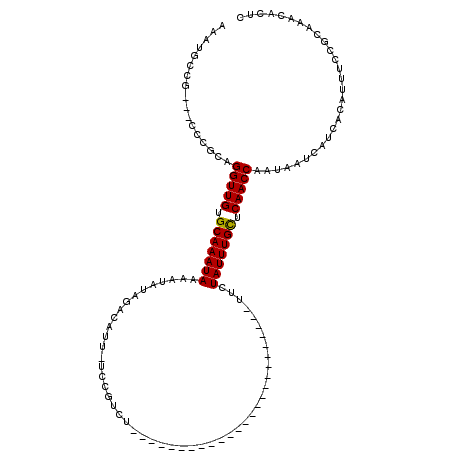
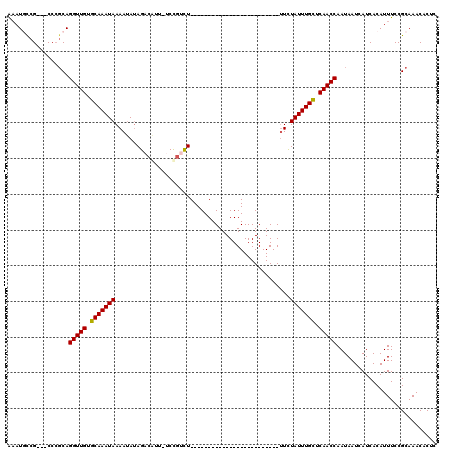
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:08 2006