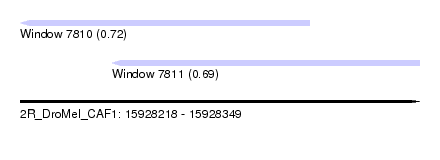
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,928,218 – 15,928,349 |
| Length | 131 |
| Max. P | 0.717625 |

| Location | 15,928,218 – 15,928,313 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -23.92 |
| Energy contribution | -25.06 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15928218 95 - 20766785 ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACA-----------CCAGG----ACACCG------GGACGCAGGUCGCUG-GACGCAGAA---C .((((((..((((((((.....)))))....(((.((((((....))))))(((((...-----------)))))----......------))))))..)))..))-)........---. ( -36.10) >DroSec_CAF1 55786 95 - 1 ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACA-----------CCAGA----ACACCG------GGACGCAGGUCGCUG-GACGCAGGA---C .((.(((..((((((((.....)))))....(((.((((((....)))))).((((...-----------)))).----......------))))))..)))((..-...)).)).---. ( -34.20) >DroSim_CAF1 54405 95 - 1 ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCGAGUACUUGCCACCUGGACA-----------CCAGA----ACACCG------GGACGCAGGUCGCUG-GACGCAGGA---C .((.(((..((((((((.....)))))....(((.((((((....)))))).((((...-----------)))).----......------))))))..)))((..-...)).)).---. ( -33.90) >DroEre_CAF1 55326 101 - 1 ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACA-----------CCAGG----ACGCCGGUCGCAGGACGCAGGACGCAG-GACGCAGGA---C .((.(((..((((((((.....)))))........((((((....))))))(((((...-----------)))))----.)))..)))((.(..(((.....)).)-..))).)).---. ( -36.60) >DroYak_CAF1 57664 94 - 1 ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUAGCAAGUACUUGCCACCUGGACA-----------CCAGG----ACG-------CAGGACGCAGGUCGCUG-GACGCAGGA---C .((.(((..((((((((.....)))))....(((...((((....))))..(((((...-----------)))))----...-------..))))))..)))((..-...)).)).---. ( -33.20) >DroPer_CAF1 57676 109 - 1 ACCCGCCACGCGGCGCUAAUAUGGCGCAACAUCCAUAGCA----UUUGCCACUUGCCGUCCCUCCUCCUCCCAAAAGAUACUCCU------GGACA-AGGUGUCCGGCACGGACGGCCGU ....(((..(..(((((.....)))))..).(((...((.----...))....(((((..(..(((..(((....((......))------)))..-))).)..))))).))).)))... ( -32.50) >consensus ACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACA___________CCAGA____ACACCG______GGACGCAGGUCGCUG_GACGCAGGA___C .((.(((..((((((((.....)))))....(((.((((((....)))))).((((..............)))).................))))))..)))((......)).))..... (-23.92 = -25.06 + 1.14)

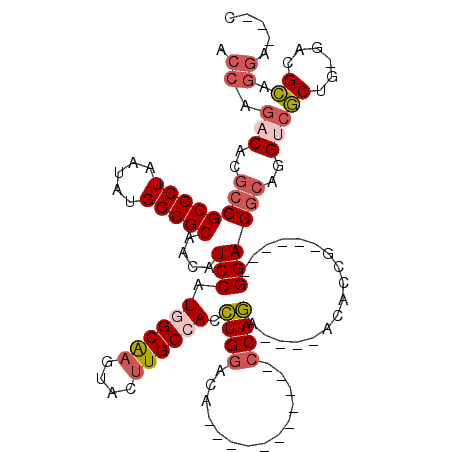
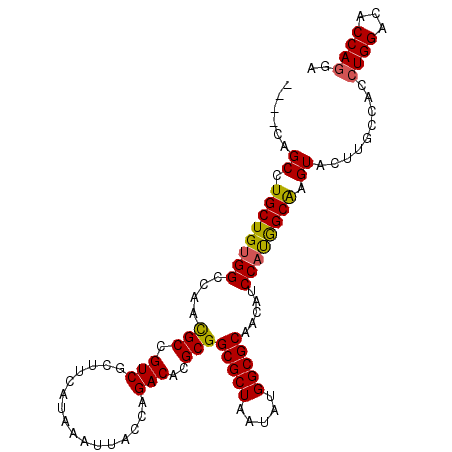
| Location | 15,928,248 – 15,928,349 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -27.13 |
| Energy contribution | -26.82 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15928248 101 - 20766785 ----CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACACCAGGA ----...((((.((((((....)))................((((....(..(((((.....)))))..).....((((((....)))))).))))))).)))). ( -34.20) >DroSec_CAF1 55816 101 - 1 ----CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACACCAGAA ----....(((.((((((....)))................((((....(..(((((.....)))))..).....((((((....)))))).))))))).))).. ( -31.00) >DroSim_CAF1 54435 101 - 1 ----CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCGAGUACUUGCCACCUGGACACCAGAA ----....(((.((((((....)))................((((....(..(((((.....)))))..).....((((((....)))))).))))))).))).. ( -30.70) >DroEre_CAF1 55362 101 - 1 ----CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACACCAGGA ----...((((.((((((....)))................((((....(..(((((.....)))))..).....((((((....)))))).))))))).)))). ( -34.20) >DroYak_CAF1 57693 101 - 1 ----CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUAGCAAGUACUUGCCACCUGGACACCAGGA ----..((.((((((((....(((.(((................)))..)))(((((.....))))).....)))))))).))........(((((...))))). ( -32.99) >DroAna_CAF1 50016 103 - 1 CGGAUAGCCUGCUGCGGCCGUUGCUGUCGCUUCAUAAAUUACCCGACACGCGGCGCUAAUAUGGCGCAACAUCCCCAGCAAGUACUUGCCACCUGGACACCAU-- .((...((.(((((.((..(((((.(((((...................)))))(((.....))))))))..)).))))).))...((((....)).))))..-- ( -30.21) >consensus ____CAGCCUGCUGUGGCCAACGCCGUCGCUUCAUAAAUUACCAGACACGCGGCGCUAAUAUGGCGCAACAUCCAUGGCAAGUACUUGCCACCUGGACACCAGGA ......((.((((((((....(((.(((................)))..)))(((((.....))))).....)))))))).)).........((((...)))).. (-27.13 = -26.82 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:06 2006