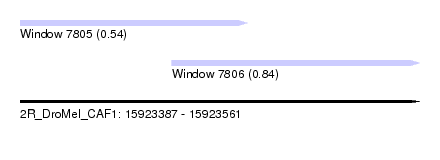
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,923,387 – 15,923,561 |
| Length | 174 |
| Max. P | 0.836745 |

| Location | 15,923,387 – 15,923,486 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
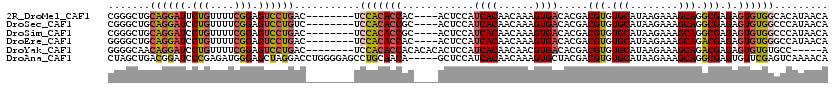
>2R_DroMel_CAF1 15923387 99 + 20766785 UAGCCGGCCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGG---GAGAGCUAGAGCUGUUGGCUUCGGGCUGCAGGAUUCUGUUUUCGGAGUCCUGAC------- (((((((((((..((((......)))).((((...(((((.....---..)))))..)))).))))))...)))))((((((((((....))))))))))..------- ( -42.80) >DroSec_CAF1 51096 99 + 1 UAUCCGGCCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGG---GAGAGCCAGGUCUGCUGGCUUCGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGUC------- ...(((((.....((((....))))....)))))((((((.....---((((((.((((((((.(.(....).).)))))..))))))))).))))))....------- ( -37.70) >DroSim_CAF1 49187 99 + 1 UAGCCGGCCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGG---GAGAGCCCCACCUGCUGGCUUCGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC------- (((((((((....((((......)))).(((..(((((((.....---..)))))))....)))))))...)))))(((((..(((....)))..)))))..------- ( -44.60) >DroEre_CAF1 50550 99 + 1 UAGCCGGUCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGG---GAGAGCCAGAGCUUCUGGCCUGGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC------- (((((........((((....)))).((.((..(((((((....(---(....)).)))))))..)).)).)))))(((((..(((....)))..)))))..------- ( -40.40) >DroYak_CAF1 52670 102 + 1 UAGCCGGCCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGGGCGGAGAGCCAGCGCUUCUGGCUUGGGGCAACAGGAUCCUGUUUUCGGAGUCCUGAC------- ..(((........((((....)))).(((((..(((((.(....(((.....))).).)))))..))))).)))..(((((..(((....)))..)))))..------- ( -43.40) >DroAna_CAF1 43958 96 + 1 UAGCCAGACGAACGUGUCAUAACACACAAGCCGGGGGCUCACUGG---GAGAGUUC----------CUUCUAGCUGACGGAUCCCGAGAUGGGAGCUAGGACCUGGGGA ...((........((((......))))...(((((..((((((((---(((.....----------))))))).))).((.((((.....)))).)).)..))))))). ( -29.20) >consensus UAGCCGGCCGAACGUGUCAUAACACACAAGCUGGGGGCUCACAGG___GAGAGCCAGAGCUGCUGGCUUCGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC_______ (((((((((....((((......))))........(((((.(......).))))).........))))...)))))((((((.(((....))).))))))......... (-26.52 = -26.58 + 0.06)

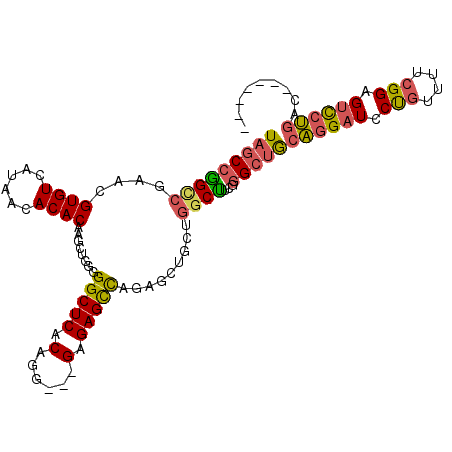
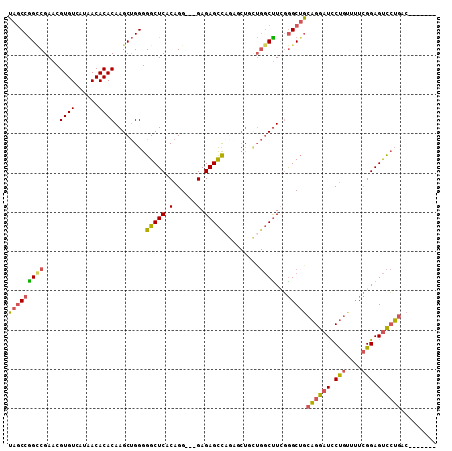
| Location | 15,923,453 – 15,923,561 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -41.05 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15923453 108 + 20766785 CGGGCUGCAGGAUUCUGUUUUCGGAGUCCUGAC--------UCCACACCAC----ACUCCAUCACAACAAAGUGACACGACGUGUGCAUAAGAAAGCAGGCGAGAGUGUGGCACAUAACA .(((...((((((((((....))))))))))..--------)))...((((----((((..((((......)))).....(((.(((........))).))).))))))))......... ( -43.70) >DroSec_CAF1 51162 108 + 1 CGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGUC--------UCCACACCGC----ACUCCAUCACAACAAAGUGACACGACGUGUGCAUAAGAAAGCAGGCGAGAGUGUGGCCCAUAACA .((((.((((((..(((....)))..)))))).--------.......(((----((((..((((......)))).....(((.(((........))).))).)))))))))))...... ( -42.90) >DroSim_CAF1 49253 108 + 1 CGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC--------UCCACACCGC----ACUCCAUCACAACAAAGUGACACGACGUGUGCAUAAGAAAGCAGGCGAGAGUGUGGCCCAUAACA .((((..((....((((((((((((((....))--------)))....(((----((((..((((......))))...)).))))).....)))))))))......))..))))...... ( -43.40) >DroEre_CAF1 50616 108 + 1 GGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC--------UCCACACCAC----ACUCCAUCACAACAAAGUGACACGACGUGUGCAUAAGAAAGCAGACGAGAGUGUGGGCCAUAACA ((((...(((((..(((....)))..))))).)--------)))...((((----((((..((((......)))).....(((.(((........))).))).))))))))......... ( -42.60) >DroYak_CAF1 52739 107 + 1 GGGGCAACAGGAUCCUGUUUUCGGAGUCCUGAC--------UCCACACCACACACACUCCAUCACAACAACGUGACACGACGUGUGCAUAAGAAAGCAGACGAGAGUGUGUGCC-----A ((((.(((((....))))).))(((((....))--------)))...)).(((((((((..((((......)))).....(((.(((........))).))).)))))))))..-----. ( -41.70) >DroAna_CAF1 44014 115 + 1 CUAGCUGACGGAUCCCGAGAUGGGAGCUAGGACCUGGGGAGCCUGCAACA-----GCUCCAUCACAACAAAGUGCUACGACGUGUGCAUAAGAAAGCAGGCGAGUGUUCGAGUCAAAACA .....((((((.((((.....)))).)).((((((..(((((........-----)))))..(((......)))......(((.(((........))).))))).))))..))))..... ( -32.00) >consensus CGGGCUGCAGGAUCCUGUUUUCGGAGUCCUGAC________UCCACACCAC____ACUCCAUCACAACAAAGUGACACGACGUGUGCAUAAGAAAGCAGGCGAGAGUGUGGCCCAUAACA .......((((((.(((....))).))))))...........(((((((............((((......)))).....(((.(((........))).))).).))))))......... (-25.02 = -25.33 + 0.31)
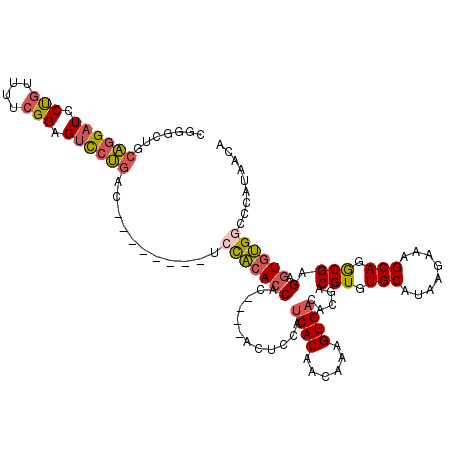
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:01 2006