
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,919,848 – 15,919,942 |
| Length | 94 |
| Max. P | 0.642611 |

| Location | 15,919,848 – 15,919,942 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -19.69 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
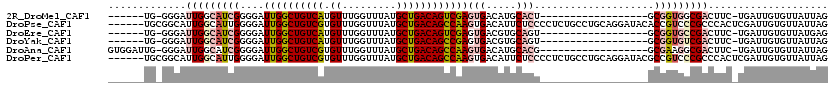
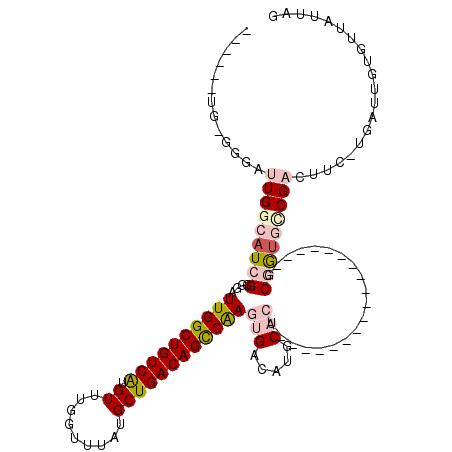
>2R_DroMel_CAF1 15919848 94 + 20766785 ------UG-GGGAUUGGCAUCGGGGAUUGGCUGUCAUGUUUGGUUUAUGCUGACAGUCGAGUGACAUGCACU------------------GCGGUGGCGACUUC-UGAUUGUGUUAUUAG ------.(-(((.(((.(((((.((.((((((((((.((.........))))))))))))((.....)).))------------------.))))).)))))))-............... ( -28.90) >DroPse_CAF1 49304 114 + 1 ------UGCGGCAUUGGCAUUGGGGAUUGGCUGUCGUGUUUGGUUUAUGCUGACAGCCAAGUGACAUUCUCCCCUCUGCCUGCAGGAUACACCGUCCCGCCCACUCGAUUGUGUUAUUAG ------.((((((..((((..((((((((((((((((((.......)))).))))))))).........)))))..))))))).((((.....))))))).(((......)))....... ( -42.10) >DroEre_CAF1 45735 94 + 1 ------UG-GGGAUUGGCAUCGGGGAUUGGCUGUCAUGUUUGGUUUAUGCUGACAGUCGAGUGACGUGCAGU------------------GCGGUGCCGACUUC-UGAUUGUGUUAUGAG ------.(-(((.(((((((((.(..((((((((((.((.........))))))))))))((.....))..)------------------.)))))))))))))-............... ( -30.30) >DroYak_CAF1 48959 94 + 1 ------UG-GGGAUUGGCAUCGGGGAUUGGCUGUCAUGUUUGGUUUAUGCUGACAGCCGAGUGACGUGCAGU------------------GCGGUGUCGACUUC-UGAUUGUGUUAUUAG ------.(-(((.(((((((((.(..((((((((((.((.........))))))))))))((.....))..)------------------.)))))))))))))-............... ( -30.30) >DroAna_CAF1 40622 100 + 1 GUGGAUUG-GGGAUUGGCAUCGGGGAUUGGCUGUCGUGUUUGGUUUAUGCUGACAGCCAAGUGACAUGCACG------------------GCGAAGGCGACUUC-UGAUUGUGUUAUUAG ...(((..-((..(((.(.(((....(((((((((((((.......)))).)))))))))(((.....))).------------------.))).).)))..))-..))).......... ( -30.40) >DroPer_CAF1 49454 114 + 1 ------UGCGGCAUUGGCAUUGGGGAUUGGCUGUCGUGUUUGGUUUAUGCUGACAGCCAAGUGACAUUCUCCCCUCUGCCUGCAGGAUACGCCGUCCCGCCCACUCGAUUGUGUUAUUAG ------.((((((..((((..((((((((((((((((((.......)))).))))))))).........)))))..))))))).((((.....))))))).(((......)))....... ( -42.10) >consensus ______UG_GGGAUUGGCAUCGGGGAUUGGCUGUCAUGUUUGGUUUAUGCUGACAGCCAAGUGACAUGCACU__________________GCGGUGCCGACUUC_UGAUUGUGUUAUUAG .............(((((((((....((((((((((.((.........))))))))))))(((.....)))....................))))))))).................... (-19.69 = -20.92 + 1.22)
| Location | 15,919,848 – 15,919,942 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.76 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
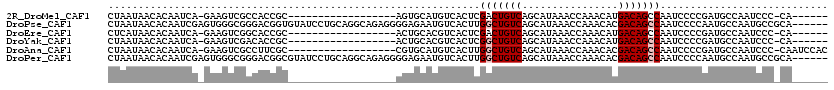
>2R_DroMel_CAF1 15919848 94 - 20766785 CUAAUAACACAAUCA-GAAGUCGCCACCGC------------------AGUGCAUGUCACUCGACUGUCAGCAUAAACCAAACAUGACAGCCAAUCCCCGAUGCCAAUCCC-CA------ ...............-(..((((....((.------------------((((.....)))))).((((((..............))))))........))))..)......-..------ ( -13.14) >DroPse_CAF1 49304 114 - 1 CUAAUAACACAAUCGAGUGGGCGGGACGGUGUAUCCUGCAGGCAGAGGGGAGAAUGUCACUUGGCUGUCAGCAUAAACCAAACACGACAGCCAAUCCCCAAUGCCAAUGCCGCA------ ................((((((((((.......)))))).((((..((((((.....)..(((((((((................))))))))))))))..))))....)))).------ ( -44.29) >DroEre_CAF1 45735 94 - 1 CUCAUAACACAAUCA-GAAGUCGGCACCGC------------------ACUGCACGUCACUCGACUGUCAGCAUAAACCAAACAUGACAGCCAAUCCCCGAUGCCAAUCCC-CA------ ...............-......((((.((.------------------...((..(((....)))(((((..............))))))).......)).))))......-..------ ( -14.14) >DroYak_CAF1 48959 94 - 1 CUAAUAACACAAUCA-GAAGUCGACACCGC------------------ACUGCACGUCACUCGGCUGUCAGCAUAAACCAAACAUGACAGCCAAUCCCCGAUGCCAAUCCC-CA------ ...........(((.-((.((.(((...((------------------...))..)))))))((((((((..............)))))))).......))).........-..------ ( -18.34) >DroAna_CAF1 40622 100 - 1 CUAAUAACACAAUCA-GAAGUCGCCUUCGC------------------CGUGCAUGUCACUUGGCUGUCAGCAUAAACCAAACACGACAGCCAAUCCCCGAUGCCAAUCCC-CAAUCCAC ...............-((((....))))..------------------.(.((((.....(((((((((................)))))))))......)))))......-........ ( -19.29) >DroPer_CAF1 49454 114 - 1 CUAAUAACACAAUCGAGUGGGCGGGACGGCGUAUCCUGCAGGCAGAGGGGAGAAUGUCACUUGGCUGUCAGCAUAAACCAAACACGACAGCCAAUCCCCAAUGCCAAUGCCGCA------ ................((((((((((.......)))))).((((..((((((.....)..(((((((((................))))))))))))))..))))....)))).------ ( -44.29) >consensus CUAAUAACACAAUCA_GAAGUCGGCACCGC__________________AGUGCAUGUCACUCGGCUGUCAGCAUAAACCAAACACGACAGCCAAUCCCCGAUGCCAAUCCC_CA______ ..............................................................(((((((................)))))))............................ ( -9.42 = -9.76 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:56 2006