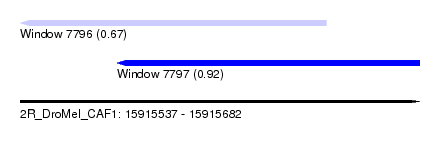
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,915,537 – 15,915,682 |
| Length | 145 |
| Max. P | 0.915890 |

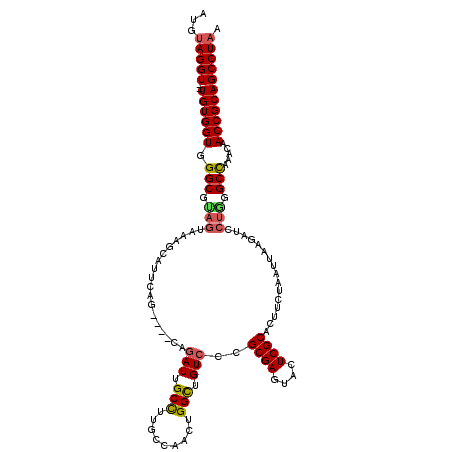
| Location | 15,915,537 – 15,915,648 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15915537 111 - 20766785 ---AUUGCCUUUCCAACUGGUUGUCCCGCGAGUACUCGCAGUUCUAAUUAAGAUCCUAGGCUAAACAACCGCAGCCUAAUCACGCUCCACCUGCCAUCUAUGCACCUUUCCACU ---..(((..........((((((.((((((....))))...(((.....))).....))....))))))((((................)))).......))).......... ( -18.79) >DroSec_CAF1 43283 114 - 1 CAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAACAUCCUGGGCCAAACAACCGCAGCCUAAUCACGCUGCACCUGCCAUCUUUGCACCUUUCCACU (((...((((.(((....)))......((((....))))..................)))).........(((((........)))))..)))..................... ( -24.40) >DroSim_CAF1 40713 114 - 1 CAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAAUCACGCUGCACCUGCCAUCUUUGCACCUUUCCACU .((((.((...(((....)))((.(((((((....))))...(((.....)))....))).)).......(((((........)))))....)).))))............... ( -27.30) >DroEre_CAF1 41464 114 - 1 UUGAUUGCUUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUGCUGGGCCAAACAACCGCAGCCUAAUCACGCUGCACCUGCCAUCUUUGCACCUUUCCACU ..........(((.((.((((((.(((((((....))))...(((.....)))....))).)).......(((((........)))))....))))..)).))).......... ( -27.10) >DroYak_CAF1 44699 114 - 1 CCGAUUGCUUCGCCAACUGGCUGUCCCGCGAGUAGUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAAUCACGCUGCACCUGCCAUCUUUGCACCUUUCCACU ..(((.((...(((....)))((.(((((((....))))...(((.....)))....))).)).......(((((........)))))....)).)))................ ( -26.50) >consensus CAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAAUCACGCUGCACCUGCCAUCUUUGCACCUUUCCACU .....(((.........((((((.(((((((....))))...(((.....)))....))).)).......(((((........)))))....)))).....))).......... (-21.04 = -21.32 + 0.28)

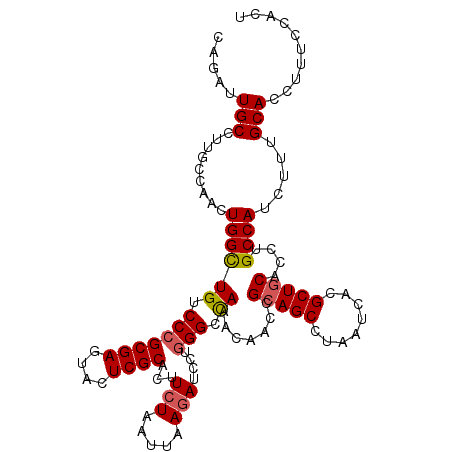
| Location | 15,915,572 – 15,915,682 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15915572 110 - 20766785 AUGUAGGU--UGUGGUGGGCGUAGUAAAGCAUUAAG-------AUUGCCUUUCCAACUGGUUGUCCCGCGAGUACUCGCAGUUCUAAUUAAGAUCCUAGGCUAAACAACCGCAGCCUAA ...(((((--((((((.(((.(((...........(-------((.(((.........))).)))..((((....))))................))).))).....))))))))))). ( -35.10) >DroSec_CAF1 43318 117 - 1 AUGUAGGU--UGUGGUGGGCGUAGUAAAGCAUUCAGAUCGCAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAACAUCCUGGGCCAAACAACCGCAGCCUAA ...(((((--((((((.(((.(((....(((..(((...((((......))))...)))..)))...((((....))))................))).))).....))))))))))). ( -40.00) >DroSim_CAF1 40748 117 - 1 AUGUAGGU--UGUGGUGGGCGUAGUAAAGCAUUCAGAUCGCAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAA ...(((((--((((((.(((.(((...........((((...(((.(((.........))).)))..((((....))))............))))))).))).....))))))))))). ( -40.10) >DroEre_CAF1 41499 110 - 1 UUGUAGGU--UGUGGUGGGCACAGUAAGGCAUU-------UUGAUUGCUUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUGCUGGGCCAAACAACCGCAGCCUAA ...(((((--((((((.(((.(.....((((((-------(((((((....(((....)))......((((....)))).....))))))))))))).)))).....))))))))))). ( -45.20) >DroYak_CAF1 44734 105 - 1 AUGCAGGUGGUGUGGUGGGCA----------UUCG----CCCGAUUGCUUCGCCAACUGGCUGUCCCGCGAGUAGUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAA .....(((((.(..((((((.----------...)----))).))..).)))))....(((((((((((((....))))...(((.....)))....)))..........))))))... ( -36.40) >consensus AUGUAGGU__UGUGGUGGGCGUAGUAAAGCAUUCAG____CAGAUUGCCUUGCCAACUGGCUGUCCCGCGAGUACUCGCACUUCUAAUUAAGAUCCUGGGCCAAACAACCGCAGCCUAA ...(((((..((((((.(((.(((..................(((.(((.........))).)))..((((....))))................))).))).....))))))))))). (-29.16 = -29.36 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:52 2006