
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,915,337 – 15,915,437 |
| Length | 100 |
| Max. P | 0.797500 |

| Location | 15,915,337 – 15,915,437 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.62 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15915337 100 + 20766785 ---------UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUGAGCUGCAAACUUGAUUAAAUGUUGCGG-----GGACGG-----CUGCUAGCCUGCUG-GCUGUCA ---------(((((((((((.....)))))))))))..((((.......((((((..((((((..((.....))..))))))-----...)))-----)))..((((....)-))))))) ( -35.80) >DroPse_CAF1 43682 95 + 1 -----CCUGUCUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCGGAGCUGCAAACUUGAUUAAAUGCUGCCG--------------------CAGCGCGCUGGGUUGUCA -----...((.(((((((((.....))))))))).)).(((((...((.(((((..(((((....................)--------------------)))).))))).))))))) ( -30.55) >DroEre_CAF1 41264 100 + 1 ---------UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUGAGCUGCAAACUUGAUUAAAUGCUGUGG-----GGAGGG-----CCGCUAGCCUGCUG-GCUGUCA ---------(((((((((((.....)))))))))))...........((((((.......((....)).......)))))).-----.((.((-----((((......)).)-))).)). ( -33.94) >DroYak_CAF1 44495 100 + 1 ---------UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUGAGCUGCAAACUUGAUUAAAUGUUGCGG-----GGAGUU-----CUGCUAGCUGGCUG-GCUGUCA ---------(((((((((((.....)))))))))))..(((((......(((((.((((((((((((...............-----.)))))-----.))).)))))))))-..))))) ( -32.39) >DroAna_CAF1 36191 110 + 1 ---------UGUCUGUUUGUUAUAGCCAAAUAGACACUUGACAUAAACACAGCUGAGCUGCAAACUUGAUUAAAUGUUGCCGCAGCGAGAUGGCGAAACGGUGAGACGACCU-CCUGUCA ---------((((((((((.......))))))))))..(((((........((((.(((((.(((..........)))...)))))....)))).....(((......))).-..))))) ( -28.10) >DroPer_CAF1 43760 100 + 1 GUCUUCCUGUCUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCGGAGCUGCAAACUUGAUUAAAUGCUGCCG--------------------CAGCGCGCUGGGUUGUCA ........((.(((((((((.....))))))))).)).(((((...((.(((((..(((((....................)--------------------)))).))))).))))))) ( -30.55) >consensus _________UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUGAGCUGCAAACUUGAUUAAAUGCUGCCG_____GGA_GG_____C_GCUAGCCCGCUG_GCUGUCA .........(((((((((((.....)))))))))))............(((((...((.(((............))).))......................(((....))).))))).. (-17.84 = -17.62 + -0.22)

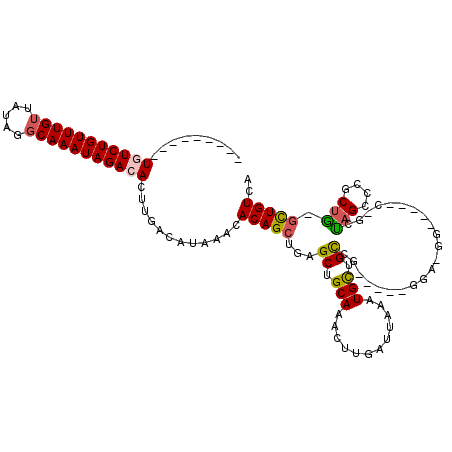
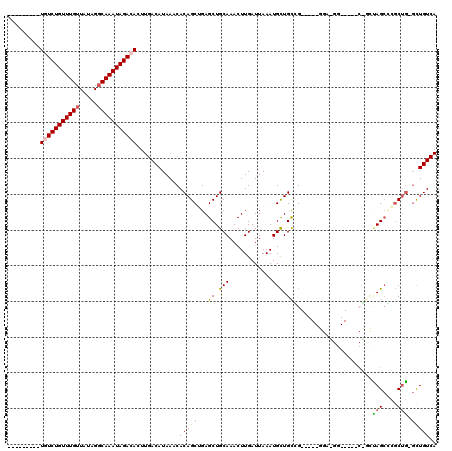
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:51 2006