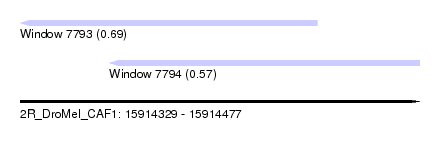
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,914,329 – 15,914,477 |
| Length | 148 |
| Max. P | 0.686415 |

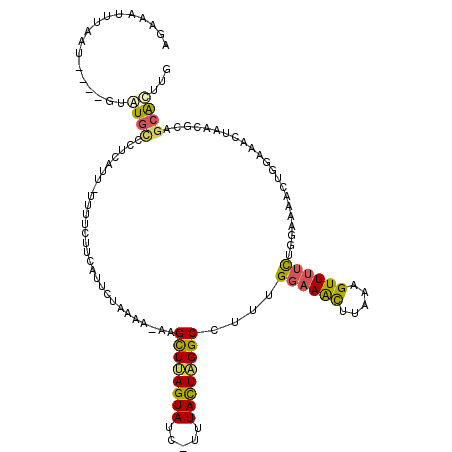
| Location | 15,914,329 – 15,914,439 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -10.90 |
| Energy contribution | -10.46 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15914329 110 - 20766785 CUAAAA-AAGCUUAGUAUC-UUUACUAGGCCUCUGCAAACUUAAAGUUUUCUUGA-AACUGGAAACUAACGCUGCACUUGUUUUGCUUCAAAAAUAUCUCUUCUCCGGCGUUC- ......-..((((((((..-..))))))))..............(((((((....-....)))))))(((((((....((((((......)))))).........))))))).- ( -23.32) >DroSec_CAF1 42097 90 - 1 CUAAAA-GAGCUUAGUAUC-UUUACUAGGCCUUUGGAAACUUAAAGUUUUCUGA---------------------GCUUGUUUUGCUACGAAAAUAUCUCUUCUCCGGCGCUC- ......-((((((((((..-..))))))(((...((((((.....)))))).((---------------------(..((((((......)))))).)))......)))))))- ( -21.80) >DroSim_CAF1 39516 111 - 1 CUUAAA-AAGCUUAGUAUA-UUUACUAGGCCUUUGGAAGCUUAAAGUUGUCUGGAAAACUUGAAACUAAGGCAGCACUUGUUUUGCUUCAAAAAUAUCUCUCUUCCGGCGCUC- ......-..((((((((..-..))))))))..(((((((......(((((((................)))))))...((((((......)))))).....))))))).....- ( -25.19) >DroEre_CAF1 40227 113 - 1 CCAAAA-ACGCUCAGUAACGUGUACUGGGCCUUUGGAAACUUAAAGUUUUCUUGAAAACUGGAAAUUAGCGUUGCAUUUGCGUUACUGCACAAAUAUCUCUUCCCCGAGGCUCA ......-..((((((((.....))))))))((((((........((((((....))))))((((....((...))(((((.((....)).))))).....)))))))))).... ( -26.20) >DroYak_CAF1 43449 114 - 1 GGAAAACACGUUCAGUAACGUGUAUUAGGCCUGUGGAAAUUUAAAGUUUUUUAGAAAACUUCAAACUGACGUAGCACUUGUGUUACUUCAAAAAUAGCAAUUCUCGGGGGCUCA .....(((((((....)))))))....(((((.(((((.(((.(((((((....))))))).))).(((.((((((....)))))).)))..........))).)).))))).. ( -29.90) >consensus CUAAAA_AAGCUUAGUAUC_UUUACUAGGCCUUUGGAAACUUAAAGUUUUCUGGAAAACUGGAAACUAACGCAGCACUUGUUUUGCUUCAAAAAUAUCUCUUCUCCGGCGCUC_ .........((((((((.....))))))))....((((((.....))))))............................................................... (-10.90 = -10.46 + -0.44)

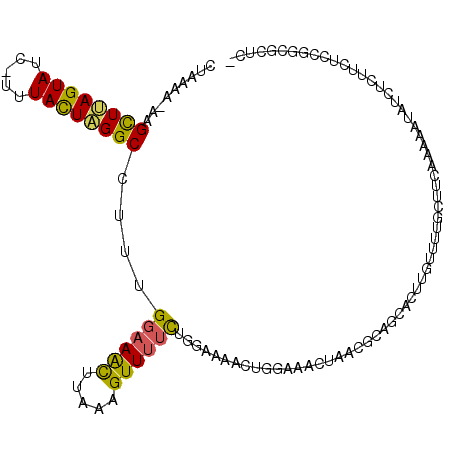
| Location | 15,914,362 – 15,914,477 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.71 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15914362 115 - 20766785 AAAAUUUUAACAUACAUAUGC-CUCAUU-UUUUCUUCAUUCUAAAA-AAGCUUAGUAUC-UUUACUAGGCCUCUGCAAACUUAAAGUUUUCUUGA-AACUGGAAACUAACGCUGCACUUG ..............((..(((-..(...-.................-..((((((((..-..))))))))..............(((((((....-....)))))))...)..)))..)) ( -18.20) >DroSec_CAF1 42130 92 - 1 GGAAAUUUAAU----GUGUGCCCUCAUU-UUUUCUUCAUUCUAAAA-GAGCUUAGUAUC-UUUACUAGGCCUUUGGAAACUUAAAGUUUUCUGA---------------------GCUUG (((((...(((----(........))))-.)))))(((.....(((-(.((((((((..-..))))))))))))((((((.....)))))))))---------------------..... ( -20.30) >DroSim_CAF1 39549 112 - 1 GGUAAUUUAAU----GUGUGCCCUCAUU-U-UUCUUCAUUCUUAAA-AAGCUUAGUAUA-UUUACUAGGCCUUUGGAAGCUUAAAGUUGUCUGGAAAACUUGAAACUAAGGCAGCACUUG ...........----(((((((......-(-(((....((((..((-(.((((((((..-..)))))))).))))))).....(((((........)))))))))....)))).)))... ( -23.10) >DroEre_CAF1 40261 115 - 1 AGCACUUUAAU----GUAUGUCUACUUCCUUCUCUUCAUUCCAAAA-ACGCUCAGUAACGUGUACUGGGCCUUUGGAAACUUAAAGUUUUCUUGAAAACUGGAAAUUAGCGUUGCAUUUG ........(((----((((((..(.((((.........(((((((.-..((((((((.....)))))))).)))))))......((((((....)))))))))).)..))).)))))).. ( -29.10) >DroYak_CAF1 43483 107 - 1 AGCAC---AAU----GUAUGUUU------UUCUCUUCAUAGGAAAACACGUUCAGUAACGUGUAUUAGGCCUGUGGAAAUUUAAAGUUUUUUAGAAAACUUCAAACUGACGUAGCACUUG ....(---(((----(((((((.------.....((((((((...(((((((....)))))))......))))))))..(((.(((((((....))))))).)))..))))).))).))) ( -23.30) >consensus AGAAAUUUAAU____GUAUGCCCUCAUU_UUUUCUUCAUUCUAAAA_AAGCUUAGUAUC_UUUACUAGGCCUUUGGAAACUUAAAGUUUUCUGGAAAACUGGAAACUAACGCAGCACUUG .................((((............................((((((((.....))))))))....((((((.....))))))......................))))... (-11.46 = -11.34 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:50 2006