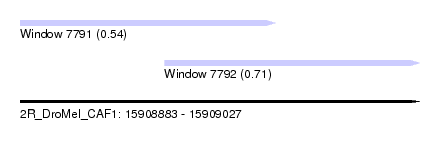
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,908,883 – 15,909,027 |
| Length | 144 |
| Max. P | 0.706590 |

| Location | 15,908,883 – 15,908,975 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -13.49 |
| Energy contribution | -14.94 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
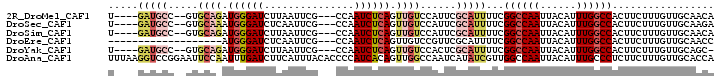
>2R_DroMel_CAF1 15908883 92 + 20766785 U----GAUGCC--GUGCAGAUGGGAUCUUAAUUCG---CCAAUCUCAGUUGUCCAUUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACA .----..(((.--((((..((((((.......)).---.((((....)))).))))..))))....((((((......))))))...........)))... ( -22.70) >DroSec_CAF1 36741 92 + 1 U----GAUGCC--GUGCAAAUGGGAUCUCAAUUCG---CCAAUCUCAGUUGUCCAUUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAAGA .----..(((.--((((.((((((....(((((..---........))))))))))).))))....((((((......))))))...........)))... ( -22.10) >DroSim_CAF1 34180 92 + 1 U----GAUGCC--GUGCAGAUGGGAUCUUAAUUCG---CCAAUCUCAGUUGUCCAUUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACA .----..(((.--((((..((((((.......)).---.((((....)))).))))..))))....((((((......))))))...........)))... ( -22.70) >DroEre_CAF1 34785 79 + 1 -------------------AUGGGAUCUCAAUUCG---CCAAUCUCAGUUGUCCGUUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACC -------------------.((((((.........---...))))))(((((..............((((((......))))))((......)).))))). ( -16.60) >DroYak_CAF1 37850 91 + 1 U----GAUGCC--GUGCAGAUGGGAUCUUAAUUCG---CCAAUCUCAGUUGUCCACUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAGC- .----(((((.--((((((.((((((.........---...)))))).)))..)))..)))))...((((((......))))))................- ( -21.70) >DroAna_CAF1 29978 101 + 1 UUUAAGGUCCGGAAUUCCAAUUUGAUCUUCAUUUACACCCCAUCACAGUUGGCCAAUCAUAUCGUUGGCCAAUUACAUUUGCCCUCUUCUUUGUUGCACCA .....(((.(((....))............................(((((((((((......))))))))))).....................).))). ( -19.80) >consensus U____GAUGCC__GUGCAGAUGGGAUCUUAAUUCG___CCAAUCUCAGUUGUCCAUUCGCAUUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACA .....(((((.....((((.((((((...............)))))).))))......)))))...((((((......))))))................. (-13.49 = -14.94 + 1.45)
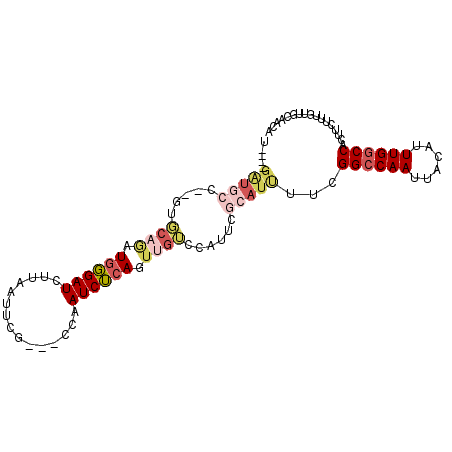
| Location | 15,908,935 – 15,909,027 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15908935 92 + 20766785 UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACAGCAAGGGCGUUGCAGGGGCACAGA----GCCA-AACAGGUGGCAGUUGGAUGGCGGU ....(.((((....((....(((((((.(((.(((((((.((....))))))))).(((.....----))))-)).)))))))...))..)))).). ( -32.40) >DroSec_CAF1 36793 78 + 1 UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAAGAGCAAGGGCGCUGCAGGGGCACAAA----GCCA-AACAUGUGGC-------------- .....((((((......))))))...((((((((.....)))))))).((..(((.(((.....----))).-..).))..))-------------- ( -26.90) >DroSim_CAF1 34232 78 + 1 UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACAGCAAGGGCGCUGCAGGGGCACAAA----GCCA-AACAUGUGGC-------------- .....((((((......))))))...(((((((((...))))))))).((..(((.(((.....----))).-..).))..))-------------- ( -28.20) >DroEre_CAF1 34824 88 + 1 UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACC-----GGCGCUGCAGGGACGCAAA----GCCAAAACAGGUGGCAGUUGGAUGGCGGC .....((((((......))))))........(((((..((-----(((..(((......)))..----((((.......)))).)))))...))))) ( -29.40) >DroYak_CAF1 37902 85 + 1 UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAGC-------GCGCUGCAGGGGCACAAC----UCCA-AACAGGUGGCAGUUGGAUGGCGGC ....(.((((....((....(((((((.(((..(((((.-------...)))))((((.....)----))))-)).)))))))...))..)))).). ( -29.60) >DroAna_CAF1 30039 84 + 1 UCGUUGGCCAAUUACAUUUGCCCUCUUCUUUGUUGCACCA---GGGGCUCUGCAGGGGCACAGGUGCCCCCA-AACAAGUGGCACU---------AU ......((((.........(((((................---)))))......((((((....))))))..-......))))...---------.. ( -25.19) >consensus UUUUCGGCCAAUUACAUUUGGCCACUUCUUUGUUGCAACA___AGGGCGCUGCAGGGGCACAAA____GCCA_AACAGGUGGCAGU_________G_ .....((((((......))))))...(((((((.((............)).)))))))..........((((.......)))).............. (-16.96 = -17.35 + 0.39)

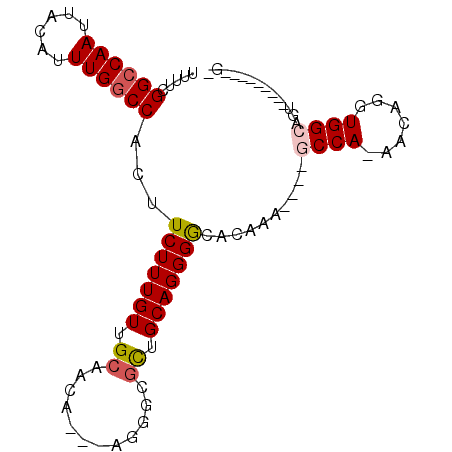
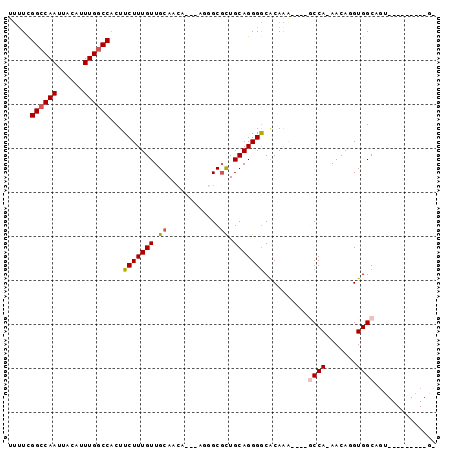
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:48 2006