| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,895,642 – 15,895,741 |
| Length | 99 |
| Max. P | 0.558190 |

| Location | 15,895,642 – 15,895,741 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
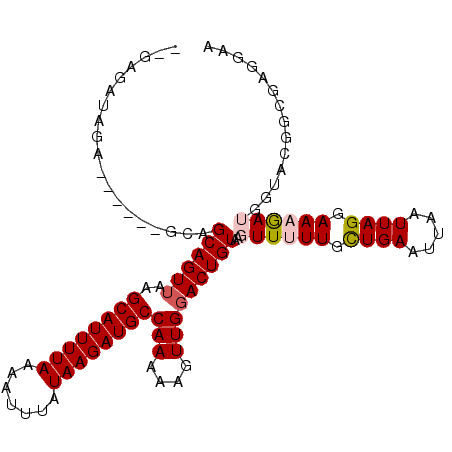
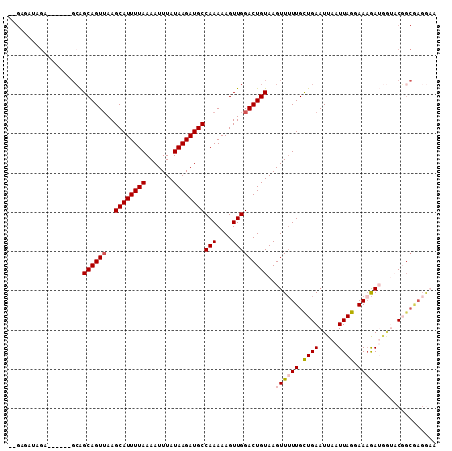
>2R_DroMel_CAF1 15895642 99 + 20766785 --G--AUAGA------GCAGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGCUGAAUUAAUUAGGAAAGAUGGUACGGCAAGGAA --.--.....------.((((..((..((((((((.......))))))))..))..))))(.(((((.((((((.((((.....)))).))))))..))))))...... ( -21.60) >DroSec_CAF1 23571 101 + 1 --GAGAUAGA------GCAGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGCUGAAUUAAUUAGGAAAGAUGGUACGGCGAGGAA --........------.((((..((..((((((((.......))))))))..))..))))(.(((((.((((((.((((.....)))).))))))..))))))...... ( -21.60) >DroSim_CAF1 21012 101 + 1 --GAGAUAGA------GCAGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGCUGAAUUAAUUAGGAAAGAUGGUACGGCGAGGAA --........------.((((..((..((((((((.......))))))))..))..))))(.(((((.((((((.((((.....)))).))))))..))))))...... ( -21.60) >DroYak_CAF1 24310 101 + 1 --GAUAGAGC------GCAGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGCUGAAUUAAUUAGGAACGAUGGAACGGCAAGGCA --........------((.((((((..((((((((.......))))))))(((....))))))))).....(((((((.....(((......)))....))))))))). ( -20.40) >DroAna_CAF1 16546 90 + 1 -----GCAAG------CCGGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGUUGAAUUAAUUAGGAAAGAGAAUUCC-------- -----(((..------(((((..((..((((((((.......))))))))..))..)))))..)))...(((((.((((.....)))).))))).......-------- ( -17.40) >DroPer_CAF1 19520 103 + 1 GAGAGAGAGAGUCGCGGGAGCAGUAAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUU-UUGUUGAAUUAAUUAGGAAAAA-----CUGCGAGAAA ...........((((((..(((((...((((((((.......))))))))(((....))).)))))...((-((.((((.....)))).)))).-----)))))).... ( -21.40) >consensus __GAGAUAGA______GCAGCAGUUAAGCAUUUUAAAAUUUAUAAGAUGCCAAAAAGUUGGACUGUAAGUUUUUGCUGAAUUAAUUAGGAAAGAUGGUACGGCGAGGAA ...................((((((..((((((((.......))))))))(((....)))))))))..((((((.((((.....)))).)))))).............. (-14.88 = -15.35 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:38 2006