| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,881,031 – 15,881,126 |
| Length | 95 |
| Max. P | 0.849670 |

| Location | 15,881,031 – 15,881,126 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
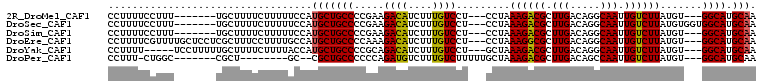
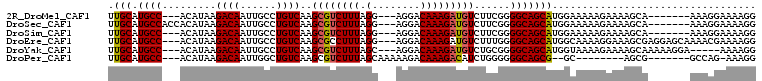
>2R_DroMel_CAF1 15881031 95 + 20766785 CCUUUUCCUUU-------UGCUUUUCUUUUUCCAUGCUGCCCCGAAGACAUCUUUGUCCU---CCUAAAGACGCUUGACAGGCAAUUGUCUUAUGU---GGCAUGCAA ...........-------................(((((((.((((((((((((((....---..)))))).(((.....)))...)))))).)).---)))).))). ( -22.00) >DroSec_CAF1 18171 98 + 1 CCUUUUCCUUU-------UGCUUUUCUUUUUCCAUGCUGCCCCGAAGACAUCUUUGUCCU---CCUAAAGACGCUUGACAGGCAAUUGUCUUAUGUGGUGGCAUGCAA ...........-------..............(((((..((.((((((((((((((....---..)))))).(((.....)))...)))))).)).))..)))))... ( -25.90) >DroSim_CAF1 15589 95 + 1 CCUUUUCCUUU-------UGCUUUUCUUUUUCCAUGCUGCCCCGAAGACAUCUUUGUCCU---CCUAAAGACGCUUGACAGGCAAUUGUCUUAUGU---GGCAUGCAA ...........-------................(((((((.((((((((((((((....---..)))))).(((.....)))...)))))).)).---)))).))). ( -22.00) >DroEre_CAF1 15916 102 + 1 CCUUUUCGUUUUGCUCCUCGCUUUCCUUUUGCCAUGCUGCCCCAAAGACAUCUUUGUCCU---CCUAAAGGCGCUUGACAGGCAAUUGUCUUAUGU---GGCAUGCAA ............((.....)).............(((((((.((((((((..(((((((.---((....)).)...))))))....)))))).)).---)))).))). ( -22.30) >DroYak_CAF1 18551 97 + 1 CCUUUU-----UCCUUUUUGCUUUUCUUUUACCAUGCUGCCCCGCAGACAUCUUUGUCCU---GCUAAAGACGCUUGACAGGCAAUUGUCUUAUGU---GGCAUGCAA ......-----.....................(((((..(...(((((((....))).))---))..((((((.(((.....))).))))))..).---.)))))... ( -24.00) >DroPer_CAF1 13017 87 + 1 CCUUU-CUGGC-------CGCU--------GC--CGCUGCCCCCCAGAUGUCUUUGUCUUUUUGCUAAAGACGCUUGACAGCCAAUUGUCUUAUGU---GGCAUGCAA ((...-..)).-------.(((--------((--(((........((.((((((((.(.....).)))))))))).(((((....)))))....))---)))).)).. ( -23.00) >consensus CCUUUUCCUUU_______UGCUUUUCUUUUUCCAUGCUGCCCCGAAGACAUCUUUGUCCU___CCUAAAGACGCUUGACAGGCAAUUGUCUUAUGU___GGCAUGCAA ..................................(((((((.....((((....)))).........((((((.(((.....))).)))))).......)))).))). (-18.28 = -18.03 + -0.25)
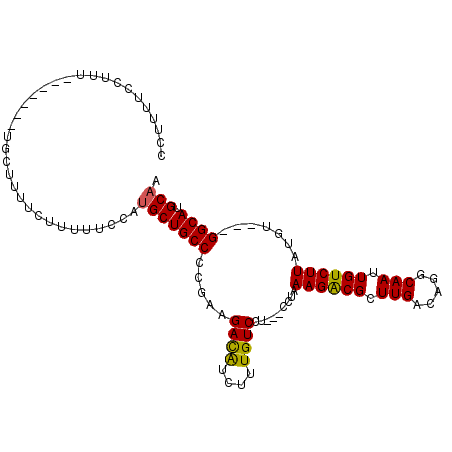
| Location | 15,881,031 – 15,881,126 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -24.32 |
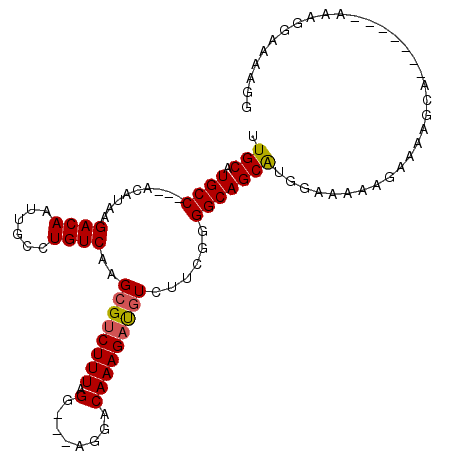
| Consensus MFE | -17.03 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15881031 95 - 20766785 UUGCAUGCC---ACAUAAGACAAUUGCCUGUCAAGCGUCUUUAGG---AGGACAAAGAUGUCUUCGGGGCAGCAUGGAAAAAGAAAAGCA-------AAAGGAAAAGG .(((.((((---....(((((..(((.....)))..)))))...(---((((((....)))))))..)))))))................-------........... ( -22.40) >DroSec_CAF1 18171 98 - 1 UUGCAUGCCACCACAUAAGACAAUUGCCUGUCAAGCGUCUUUAGG---AGGACAAAGAUGUCUUCGGGGCAGCAUGGAAAAAGAAAAGCA-------AAAGGAAAAGG .(((.((((.((....(((((..(((.....)))..)))))...(---((((((....))))))))))))))))................-------........... ( -25.50) >DroSim_CAF1 15589 95 - 1 UUGCAUGCC---ACAUAAGACAAUUGCCUGUCAAGCGUCUUUAGG---AGGACAAAGAUGUCUUCGGGGCAGCAUGGAAAAAGAAAAGCA-------AAAGGAAAAGG .(((.((((---....(((((..(((.....)))..)))))...(---((((((....)))))))..)))))))................-------........... ( -22.40) >DroEre_CAF1 15916 102 - 1 UUGCAUGCC---ACAUAAGACAAUUGCCUGUCAAGCGCCUUUAGG---AGGACAAAGAUGUCUUUGGGGCAGCAUGGCAAAAGGAAAGCGAGGAGCAAAACGAAAAGG .....((((---(.....((((......))))..(((((((...(---((((((....)))))))))))).)).)))))........((.....))............ ( -27.20) >DroYak_CAF1 18551 97 - 1 UUGCAUGCC---ACAUAAGACAAUUGCCUGUCAAGCGUCUUUAGC---AGGACAAAGAUGUCUGCGGGGCAGCAUGGUAAAAGAAAAGCAAAAAGGA-----AAAAGG .(((.((((---.(...(((((.(((((((..(((....)))..)---))).)))...)))))...)))))))).......................-----...... ( -23.80) >DroPer_CAF1 13017 87 - 1 UUGCAUGCC---ACAUAAGACAAUUGGCUGUCAAGCGUCUUUAGCAAAAAGACAAAGACAUCUGGGGGGCAGCG--GC--------AGCG-------GCCAG-AAAGG ..((.((((---.(...(((......(((....)))((((((..(.....)..)))))).)))...)))))))(--((--------....-------)))..-..... ( -24.60) >consensus UUGCAUGCC___ACAUAAGACAAUUGCCUGUCAAGCGUCUUUAGG___AGGACAAAGAUGUCUUCGGGGCAGCAUGGAAAAAGAAAAGCA_______AAAGGAAAAGG .(((.((((.........((((......))))..((((((((.(........)))))))))......))))))).................................. (-17.03 = -17.08 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:32 2006