
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,879,593 – 15,879,859 |
| Length | 266 |
| Max. P | 0.754215 |

| Location | 15,879,593 – 15,879,691 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15879593 98 + 20766785 -GGAA-------UCGCCGGCAAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCU--------------CGGCCAU -....-------..((((((...((((.((((((.(((...)))...))))))...)))).....(((((.((((((.....)))))))))))...)).--------------))))... ( -30.80) >DroPse_CAF1 11436 97 + 1 GGAAA-------UCGACGGCAAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACGGCCGCGAGCUGUCACUGCUGUCC----------------AGCAUA .....-------..(((((((..((((.((((((.(((...)))...))))))...))))..........(((((((.....))))))).))))))).----------------...... ( -25.60) >DroEre_CAF1 14480 98 + 1 -CGAA-------UCGGCGGCCAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCU--------------CGGCCAU -....-------.....((((..((((.((((((.(((...)))...))))))...)))).....((((((((((((.....))))))))....)))).--------------.)))).. ( -31.00) >DroYak_CAF1 17100 98 + 1 -CGAA-------GCGACGGCGAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCU--------------CGGCCAU -(((.-------(((.((((((.((((.((((((.(((...)))...))))))...)))).))......((((((((.....)))))))).))))))))--------------))..... ( -32.60) >DroAna_CAF1 9687 119 + 1 -CUGGUUCCUGUCCCCUGGUAAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACAGCCGCGAGCUGUCAAUGCUGCGUUCGGACAGGACCAUUCGGCCAU -.((((.(((((((.........((((.((((((.(((...)))...))))))...))))...((((((((((((((.....)))))).)))).))))..)))))))))))......... ( -41.10) >DroPer_CAF1 11471 97 + 1 GGAAA-------UCGACGGCAAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACGGCCGCGAGCUGUCACUGCUGUCC----------------AGCAUA .....-------..(((((((..((((.((((((.(((...)))...))))))...))))..........(((((((.....))))))).))))))).----------------...... ( -25.60) >consensus _CGAA_______UCGACGGCAAAUGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACAGCCGCGAGCUGUCAAUGCUGCGCU______________CGGCCAU .................(((...((((.((((((.(((...)))...))))))...)))).....((((.(((((((.....)))))))))))......................))).. (-22.39 = -22.70 + 0.31)

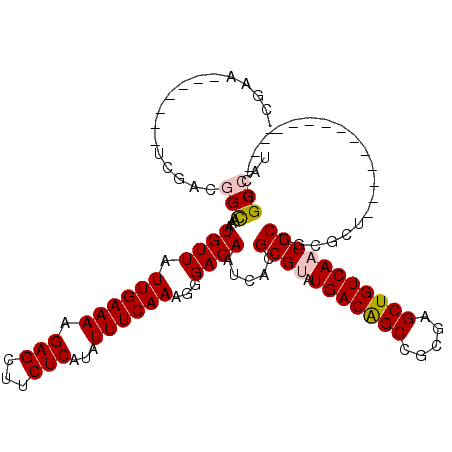
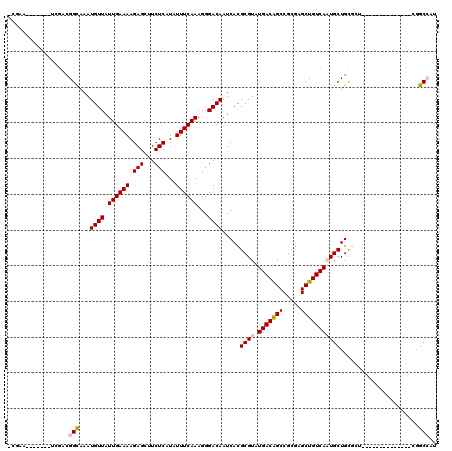
| Location | 15,879,665 – 15,879,756 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15879665 91 + 20766785 GAGCUGUCAAUGCUGCGCUCGGCCAUUCGCUCGACUAUAUAGUAUAUAUAUAUAU---------------AUAUCUCUGGCCGAUCCGGCCGGUGGCCAGAUACUG ((((.((.......))))))(((((................(((((((....)))---------------))))..(((((((...))))))))))))........ ( -27.60) >DroSec_CAF1 16813 89 + 1 GAGCUGUCAAUGCUGCGCUCGGCCAUUCGCUCGACUAUAUAGUGUAUAUAUAUA-----------------UAUAUCUGGCCGAUCGGGCUGGUGGCCAGAUACUG ((((.((.......))))))((((((..((((((.....(((((((((....))-----------------)))).))).....))))))..))))))........ ( -31.30) >DroSim_CAF1 14202 89 + 1 GAGCUGUCAAUGCUGCGCUCGGCCAUUCGCUCGACUAUAUAGUAUAUAUAUAUA-----------------UAUCUCUGGCCGAUCGGGCUGUUGGCCAGAUACUG ((((.((.......))))))(((((...((((((.....(((.(((((....))-----------------)))..))).....))))))...)))))........ ( -26.40) >DroEre_CAF1 14552 106 + 1 GAGCUGUCAAUGCUGCGCUCGGCCAUUCCCUCGACUAUAUAGUACAUACAUAUAUAGCGUAGCAUAGGGCAUAUCUCUGGCCGAUCGGGCUGUUGGCCAGAUACUG ((..((((.(((((((((((((........)))).((((((((....)).)))))))))))))))..))))..))((((((((((......))))))))))..... ( -38.30) >DroYak_CAF1 17172 85 + 1 GAGCUGUCAAUGCUGCGCUCGGCCAUUCCCUUGACUAUAUAGUAUAUACAUAUA---------------------UCUGGCCGAUCGGUCUGGUGGCCAGAUAGCA ..((((((........(.(((((((.......((.(((((.((....)))))))---------------------))))))))).)((((....)))).)))))). ( -27.81) >consensus GAGCUGUCAAUGCUGCGCUCGGCCAUUCGCUCGACUAUAUAGUAUAUAUAUAUA_________________UAUCUCUGGCCGAUCGGGCUGGUGGCCAGAUACUG ((((.((.......))))))(((((((.((((((.((((((.....)))))).......................((.....)))))))).)))))))........ (-21.02 = -22.50 + 1.48)

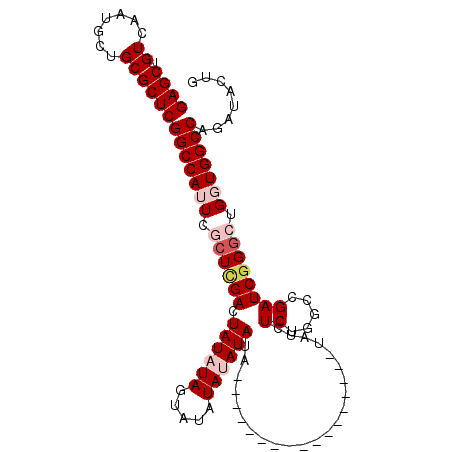
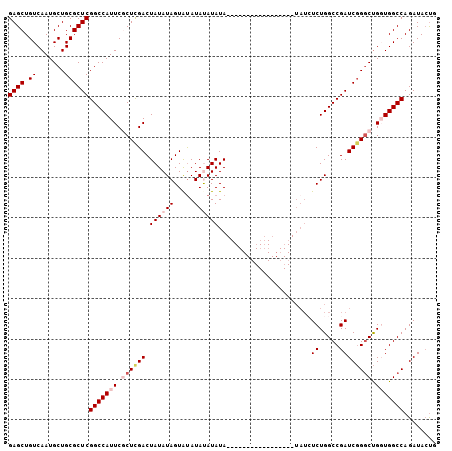
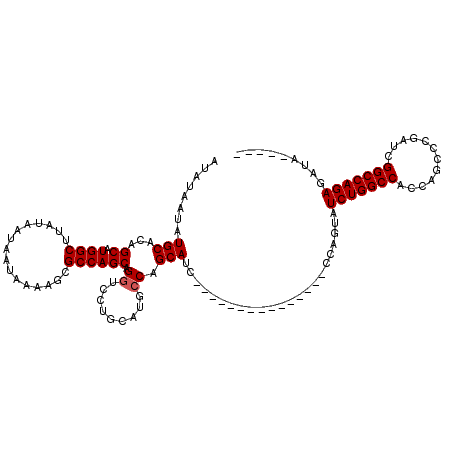
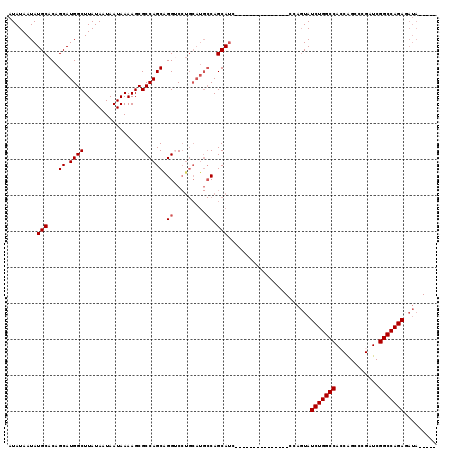
| Location | 15,879,720 – 15,879,820 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15879720 100 - 20766785 AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUCCUGCAUGCCAGCAAC---------------CCAGUAUCUGGCCACCGGCCGGAUCGGCCAGAGAUAU---- ........(((...(((((((((.((....)).))))(((....)))....)))))..)))..---------------......(((((((.((....))...))))))).....---- ( -27.60) >DroSec_CAF1 16867 91 - 1 AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUC--------CAGCAUC---------------CCAGUAUCUGGCCACCAGCCCGAUCGGCCAGAUAUA----- .......((((((.((.((((................))))))..)).--------..)))).---------------...((((((((((............)))))))))).----- ( -23.89) >DroSim_CAF1 14256 99 - 1 AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUCCUGCAUGCCAGCAUC---------------CCAGUAUCUGGCCAACAGCCCGAUCGGCCAGAGAUA----- .......((((...(((((((((.((....)).))))(((....)))....)))))..)))).---------------......(((((((............)))))))....----- ( -25.70) >DroEre_CAF1 14618 119 - 1 AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGGCCUGCAUGCCAGCAUCCCAGCAUCCAGCAUCCCAGUAUCUGGCCAACAGCCCGAUCGGCCAGAGAUAUGCCC .......((((...((.((((................)))))).(((..(((......))).))).))))...((((.......(((((((............)))))))...)))).. ( -31.79) >DroYak_CAF1 17226 95 - 1 AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCGGGGCCUGCAUGCCAGCAUC---------------CUGCUAUCUGGCCACCAGACCGAUCGGCCAGA--------- ......(((((...)))))((((.((....)).))))...(((((((..(((......)))))---------------))))).(((((((............)))))))--------- ( -28.20) >consensus AUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUCCUGCAUGCCAGCAUC_______________CCAGUAUCUGGCCACCAGCCCGAUCGGCCAGAGAUA_____ ........(((...((.((((................)))))).((.........)).))).......................(((((((............)))))))......... (-21.17 = -21.37 + 0.20)

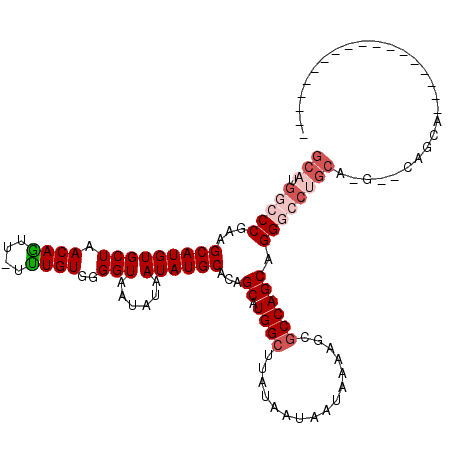
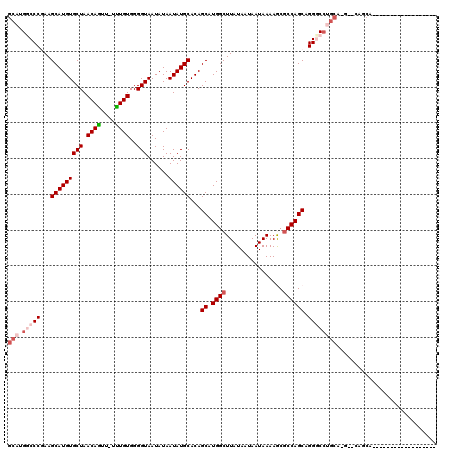
| Location | 15,879,756 – 15,879,859 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -22.72 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15879756 103 - 20766785 GCAUGGCCCGAAGCAUGUGCUAACAGUU-UUUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUCCUGCAUG-CCAGCAAC---------------C ((.((((....((.((.((((....(((-(((((...((.(((...(((((...)))))...))).)).))))))))...)))).)).))....)-)))))...---------------. ( -27.10) >DroPse_CAF1 11577 101 - 1 GCAUGCCCCAAAGCAUGUGCUAACAGUU-UCUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAACGCCAGCAGGGCCUGCAGCAGCAGCA------------------ (((.((((....(((((((((.((((..-.))))..)))......))))))...((.((((................)))))).)))).))).((....)).------------------ ( -29.39) >DroSec_CAF1 16902 96 - 1 GCAUGGCCCGAAGCAUGUGCUAACAAUUUUUUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGUC---------CAGCAUC---------------C ((.(((.((...(((((((((.((((....))))..)))......))))))...((.((((................)))))).)).)---------))))...---------------. ( -24.69) >DroEre_CAF1 14658 118 - 1 GCAUGGCCCGAAGCAUGUGCUAACAGUU-UUUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGGCCUGCAUG-CCAGCAUCCCAGCAUCCAGCAUCC (((.((((((((((.(((....))))))-))(((..(((.......(((((...)))))((((.((....)).))))))).)))))))))))(((-(..((......)).....)))).. ( -32.50) >DroAna_CAF1 9849 92 - 1 GCCUGGCCCGAAGCAUGUGCUAACAGUU-UUUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCCCCAGCAGGGCCUGCA--------------------------- ((..((((((((((.(((....))))))-))((((((((.......(((((...)))))..((((....))))..)))))).))))))).)).--------------------------- ( -30.50) >DroPer_CAF1 11612 101 - 1 GCAUGCCCCAAAGCAUGUGCUAACAGUU-UCUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAACGCCAGCAGGGCCUGCAGGAGCAGCA------------------ ((((((......))))))(((....(((-((((..(((((........)))...((.((((................))))))....))..)))))))))).------------------ ( -31.49) >consensus GCAUGGCCCGAAGCAUGUGCUAACAGUU_UUUGUGGGGUAAUAUAAUAUGCACAGCAUGGCUUAUAAUAAUAAAAGCGCCAGCAGGGCCUGCA_G__CAGCA__________________ (((.(((((...(((((((((.((((....))))..)))......))))))...((.((((................)))))).))))))))............................ (-22.72 = -24.02 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:30 2006