
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,878,983 – 15,879,076 |
| Length | 93 |
| Max. P | 0.603136 |

| Location | 15,878,983 – 15,879,076 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
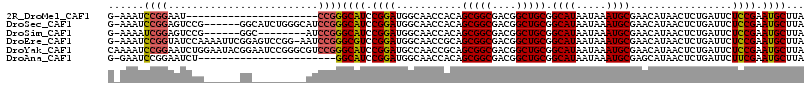
>2R_DroMel_CAF1 15878983 93 + 20766785 G-AAAUCCGGAAU----------------------CCGGGCAUCCGGAUGGCAACCACAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA .-....((((...----------------------))))((((.((((..(((......(((((....))))).((((.....))))...........)).)..)))).))))... ( -26.80) >DroSec_CAF1 16146 109 + 1 G-AAAUCCGGAGUCCG------GGCAUCUGGGCAUCCGGGCAUCCGGAUGGCAACCACAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA .-....((((((((((------(....)))))).)))))((((.((((..(((......(((((....))))).((((.....))))...........)).)..)))).))))... ( -39.60) >DroSim_CAF1 13570 101 + 1 G-AAAAUCGGAGUCCG------GGC--------AUCCGGGCAUCCGGAUGGCAACCACAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA (-(.(((((((((...------(.(--------((((((....))))))).).......(((((....))))).((((.....)))).......))))))))).)).......... ( -36.40) >DroEre_CAF1 13853 114 + 1 G-AAAUCCGGUAUCCAAAAUUCGGAGUCCGG-AAUCCGGGCGUCCGGAUGGCAACCGCAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA .-...(((((..(((.......)))..))))-)....((((((.((((..(((..(((((((....).))))))((((.....))))...........)).)..)))).)))))). ( -38.90) >DroYak_CAF1 16409 116 + 1 CAAAAUCCGGAAUCUGGAAUACGGAAUCCGGGCGUCCGGGCAUCCGGAUGCCAACCGCAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA .......((((.((((.....)))).))))(((((((((....)))))))))...(((((((....).))))))((((.....))))(((((......)).)))............ ( -44.20) >DroAna_CAF1 9327 92 + 1 G-GAAUCCGGAAUCU-----------------------GGCAUCCGGAUGGCAACCACAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAGCAUAACUCUGAUUCUUCGAAUGCUUA (-(.(((((((....-----------------------....))))))).....))...(((((....))))).((((.....))))((((((...((.((....)))))))))). ( -27.60) >consensus G_AAAUCCGGAAUCCG______GG_________AUCCGGGCAUCCGGAUGGCAACCACAGCGGCGACGGCUGCGGCAUAAUAAAUGCGAACAUAACUCUGAUUCUCCGAAUGCUUA ......((((.........................))))((((.((((...........(((((....))))).((((.....)))).................)))).))))... (-23.56 = -23.64 + 0.09)
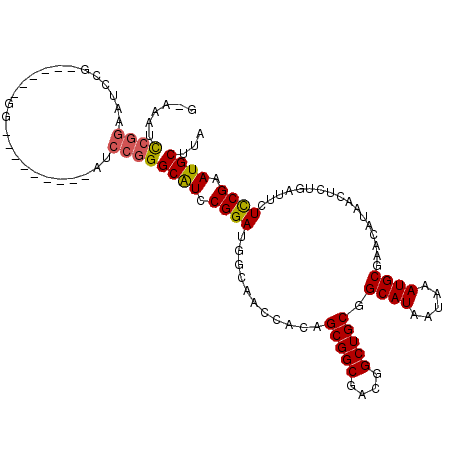

| Location | 15,878,983 – 15,879,076 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -27.57 |
| Energy contribution | -28.68 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
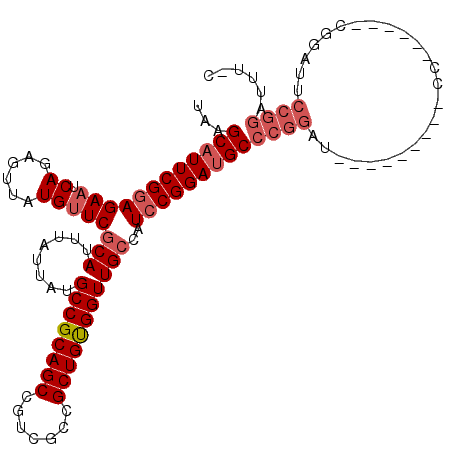
>2R_DroMel_CAF1 15878983 93 - 20766785 UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGG----------------------AUUCCGGAUUU-C ...((((((((((((.((......)))))(((........((((((((.......)))))))))))..)))))))))((((----------------------...))))....-. ( -33.80) >DroSec_CAF1 16146 109 - 1 UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAUGCCCAGAUGCC------CGGACUCCGGAUUU-C .....((((((((.....(((.....)))((((((.....((((((((.......))))))))...(((((((....)))))))...)))))).------....))))))))..-. ( -39.50) >DroSim_CAF1 13570 101 - 1 UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAU--------GCC------CGGACUCCGAUUUU-C ..........(((((((.(((((......(((........((((((((.......)))))))))))(((((((....))))))--------)..------..))))).))))))-) ( -36.70) >DroEre_CAF1 13853 114 - 1 UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGCGGUUGCCAUCCGGACGCCCGGAUU-CCGGACUCCGAAUUUUGGAUACCGGAUUU-C ...((.(((((((((.((......)))))(((........((((((((.......)))))))))))..)))))).))..(((.(-((((..(((((...)))))..))))).))-) ( -39.80) >DroYak_CAF1 16409 116 - 1 UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGCGGUUGGCAUCCGGAUGCCCGGACGCCCGGAUUCCGUAUUCCAGAUUCCGGAUUUUG ...((((((((((((.((......)))))((.........((((((((.......))))))))..)).)))))))))(((((..(..(((........))).)..)))))...... ( -41.90) >DroAna_CAF1 9327 92 - 1 UAAGCAUUCGAAGAAUCAGAGUUAUGCUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCC-----------------------AGAUUCCGGAUUC-C ...(((............(((.....)))...........((((((((.......))))))))))).(((((((....-----------------------....)))))))..-. ( -27.60) >consensus UAAGCAUUCGGAGAAUCAGAGUUAUGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAU_________CC______CGGAUUCCGGAUUU_C ...((((((((((((.((......)))))(((........((((((((.......)))))))))))..)))))))))((((.........................))))...... (-27.57 = -28.68 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:26 2006