| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,874,247 – 15,874,404 |
| Length | 157 |
| Max. P | 0.665596 |

| Location | 15,874,247 – 15,874,364 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -15.72 |
| Energy contribution | -17.19 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
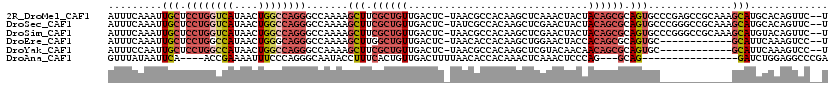
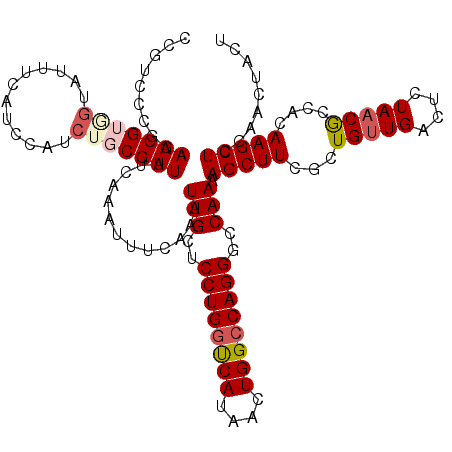
>2R_DroMel_CAF1 15874247 117 - 20766785 AUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUC-UAACGCCACAAGCUCAAACUACUACAGCGCAGUGCCCGAGCCGCAAAGCAUGCACAGUUC--U ..........((((.((((((....))))))((((.....(((.((((((......-....((.....))..........)))))).))))))))))).(((.....))).......--. ( -32.65) >DroSec_CAF1 11521 117 - 1 AUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUC-UAUCGCCACAAGCUCGAACUACUACAGCGCAGUGCCCGGGCCGCAAAGCAUGCACAGUUC--U ........((((..(((((((....)))))))((((....(((.((((((......-....((.....))..........)))))).))).....))))))))..............--. ( -35.35) >DroSim_CAF1 8966 117 - 1 AUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUC-UAACGCCACAAGCUCGAACUACUACAGCGCAGUGCCCGGGCCGCAAAGCAUGUACAGUUC--U ........((((..(((((((....)))))))((((....(((.((((((......-....((.....))..........)))))).))).....))))))))..............--. ( -35.35) >DroEre_CAF1 9407 105 - 1 AUUUCAAAUUGCUCCUGGCCAUAACUGGGCAGGGCCAAAAGCUUGGCUGUUGACUC-UAACACCACAAGCUGGAACUACCACAGCGCAGUGC------------GCAUUCAAAGUCC--U ........(((..((((.(((....))).))))..))).(((((((.(((((....-))))).).))))))(((.((......((((...))------------))......)))))--. ( -31.90) >DroYak_CAF1 11913 105 - 1 AUUUCCAAUUGCUCCUGGCCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUC-UAACGCCACAAGCUCGUACAACAACAGCGCAGUGC------------GCAUUCAAAGUCC--U .........((((((((((((....)))))))))......(((.((((((((....-....((.....))........)))))))).)))..------------)))..........--. ( -33.59) >DroAna_CAF1 5090 97 - 1 GUUUAUAAUUCA----ACCGAAAAUUUCCCAGGGCAAUACCUUUCACUGUUGACUUUUAACACCACAAACUCAAACUCCCAG---GCAG----------------GAUCUGGAGGCCCGA ............----...............((((............((((((...)))))).............((((.((---(...----------------..))))))))))).. ( -16.40) >consensus AUUUCAAAUUGCUCCUGGCCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUC_UAACGCCACAAGCUCGAACUACUACAGCGCAGUGC____________GCAUGCACAGUCC__U .........(((.((((((((....)))))))).......(((.((((((..............................)))))).)))..............)))............. (-15.72 = -17.19 + 1.48)
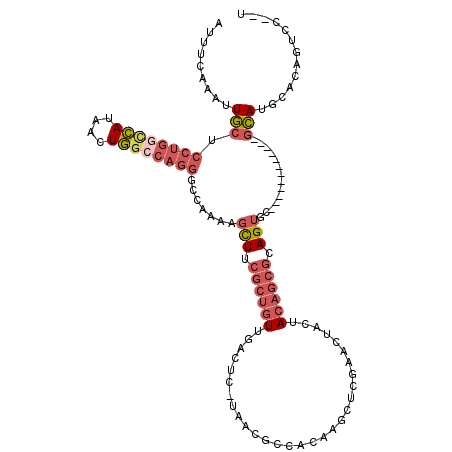
| Location | 15,874,285 – 15,874,404 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15874285 119 - 20766785 CCGUUCUGGAACGUGGUAUUUCAUCCAUCAGCGUUAUCAAAUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUCUAACGCCACAAGCUCAAACUACU ..(((..((...((((..((.....((.(((((....................((((((((....))))))))((.....))..))))).)).....))..))))...))..))).... ( -31.30) >DroSec_CAF1 11559 119 - 1 CCGUCCCGGAACGUGGUAUUUCAUCCAUCUGCGUUAUCAAAUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUCUAUCGCCACAAGCUCGAACUACU (((...)))((((..(............)..)))).......(((...(((..((((((((....))))))))..))).(((((.(.((.(((.....))).)))))))).)))..... ( -27.40) >DroSim_CAF1 9004 119 - 1 CCGUCCCGAAACGUGGUAUUUCAUCCAUCUGCGUUAUCAAAUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUCUAACGCCACAAGCUCGAACUACU ......(((...((((........))))..((((((..............((.((((((((....))))))))))(((.(((...))).)))....)))))).......)))....... ( -29.00) >DroEre_CAF1 9433 119 - 1 CCGUCCCGCAACGUUAUAUUUCAAACAUCUCCGUUAUCAAAUUUCAAAUUGCUCCUGGCCAUAACUGGGCAGGGCCAAAAGCUUGGCUGUUGACUCUAACACCACAAGCUGGAACUACC .(((......)))................(((................(((..((((.(((....))).))))..))).(((((((.(((((....))))).).)))))))))...... ( -27.90) >DroYak_CAF1 11939 119 - 1 CCGUCCCGGAACGCAGUAUUUCAUACAGCUCCGUUAUCAAAUUUCCAAUUGCUCCUGGCCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUCUAACGCCACAAGCUCGUACAACA ......((((..((.((((...)))).))))))...............(((..((((((((....))))))))..))).(((((.((.((((....))))))...)))))......... ( -30.80) >consensus CCGUCCCGGAACGUGGUAUUUCAUCCAUCUGCGUUAUCAAAUUUCAAAUUGCUCCUGGUCAUAACUGGCCAGGGCCAAAAGCUUCGCUGUUGACUCUAACGCCACAAGCUCGAACUACU .........(((((((............))))))).............(((..((((((((....))))))))..))).(((((...(((((....)))))....)))))......... (-22.20 = -23.12 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:21 2006