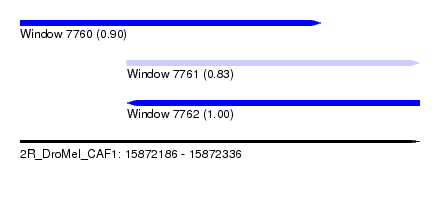
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,872,186 – 15,872,336 |
| Length | 150 |
| Max. P | 0.998724 |

| Location | 15,872,186 – 15,872,299 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
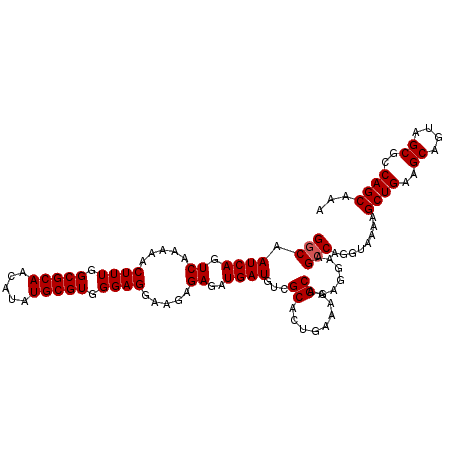
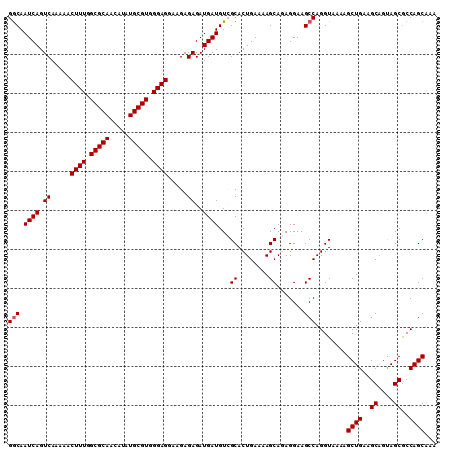
>2R_DroMel_CAF1 15872186 113 + 20766785 CUCCCUUAAGCGCAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGCGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCC .........((((((((((((..........((((.((.....))))))((....))))))))))......(((.......))).....)))).................... ( -22.00) >DroSec_CAF1 9495 113 + 1 CUCCCUUAAGCGAAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGUGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCC ........(((....)))............((((..(((....(((((.((....)).)))))........(((.......)))....)))..))))................ ( -19.70) >DroSim_CAF1 6930 113 + 1 CUCCCUUAAGCGCAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGCGCUGCUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCC .........((((((((((((..........((((.((.....))))))((....))))))))))......(((.......))).....)))).................... ( -22.00) >DroEre_CAF1 7351 113 + 1 CUCCCUUAAGCGCAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGCGCUACUGCUUCAGCAUUUACCUGGCUUCCUUUGCUUUUCAGUGCCACAUCAUCUCUCUUCCUCC .........((((..((((((..........((((.((.....))))))((....))))))))........(((.......))).....)))).................... ( -20.10) >DroYak_CAF1 9774 113 + 1 CUCCCUUAAGCGCAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGCGCUACAGCUUCAGCAUUUACCUGGCUUCCUUUGCUUUUCAGUGCCACAUCAUCUCUCUUCCUCC .........((((..((((((..........((((.((.....))))))((....))))))))........(((.......))).....)))).................... ( -19.80) >consensus CUCCCUUAAGCGCAAGUUGAACAUUUCAUUUGUCAUGCUUUUUGCUGGCGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCC .........((((..((((((..........((((.((.....))))))((....))))))))........(((.......))).....)))).................... (-18.18 = -18.22 + 0.04)

| Location | 15,872,226 – 15,872,336 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -21.75 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15872226 110 + 20766785 UUUGCUGGCGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGCC ...(((((.((....)).)))))........(((.((.((.(((((...(((((((((......................))))))))).)))))....))..))..))) ( -23.95) >DroSec_CAF1 9535 110 + 1 UUUGCUGGUGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGCC ...(((((.((....)).)))))........(((.((.((.(((((...(((((((((......................))))))))).)))))....))..))..))) ( -23.95) >DroSim_CAF1 6970 110 + 1 UUUGCUGGCGCUGCUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGCC ...(((((.((....)).)))))........(((.((.((.(((((...(((((((((......................))))))))).)))))....))..))..))) ( -23.95) >DroEre_CAF1 7391 110 + 1 UUUGCUGGCGCUACUGCUUCAGCAUUUACCUGGCUUCCUUUGCUUUUCAGUGCCACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGCC ..((((((.((....)).)))))).................((...(((((........................((((.....)))).(((.....))))))))..)). ( -21.00) >DroYak_CAF1 9814 110 + 1 UUUGCUGGCGCUACAGCUUCAGCAUUUACCUGGCUUCCUUUGCUUUUCAGUGCCACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGUC ...((((......)))).((((((...((.((((.......((........)).......................(((.....)))))))..))...)).))))..... ( -20.80) >consensus UUUGCUGGCGCUACUGCUUCAGCUUUUACCUGGCUUCCUCUGCUUUUCAGUGCGACAUCAUCUCUCUUCCUCCCACGCAUAUGUUGCGCCAAAGUUUUUGACUGAUUGCC ...(((((.((....)).)))))........(((.((.((.(((((...(((((((((......................))))))))).)))))....))..))..))) (-21.75 = -21.75 + 0.00)


| Location | 15,872,226 – 15,872,336 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15872226 110 - 20766785 GGCAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUCGCACUGAAAAGCAGAGGAAGCCAGGUAAAAGCUGAAGCAGUAGCGCCAGCAAA (((.((((.((.....((((.(((((.....))))).)))).....))..)))).((.(.(((.....))).))).)))........((((..((....))..))))... ( -29.90) >DroSec_CAF1 9535 110 - 1 GGCAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUCGCACUGAAAAGCAGAGGAAGCCAGGUAAAAGCUGAAGCAGUAGCACCAGCAAA (((.((((.((.....((((.(((((.....))))).)))).....))..)))).((.(.(((.....))).))).)))........((((..((....))..))))... ( -29.50) >DroSim_CAF1 6970 110 - 1 GGCAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUCGCACUGAAAAGCAGAGGAAGCCAGGUAAAAGCUGAAGCAGCAGCGCCAGCAAA (((.((((.((.....((((.(((((.....))))).)))).....))..)))).((.(.(((.....))).))).)))........((((..((....))..))))... ( -29.90) >DroEre_CAF1 7391 110 - 1 GGCAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUGGCACUGAAAAGCAAAGGAAGCCAGGUAAAUGCUGAAGCAGUAGCGCCAGCAAA (((..(((((......((((.(((((.....))))).))))...........((.((((.((.........))...)))).))....))))).((....)))))...... ( -29.80) >DroYak_CAF1 9814 110 - 1 GACAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUGGCACUGAAAAGCAAAGGAAGCCAGGUAAAUGCUGAAGCUGUAGCGCCAGCAAA (((.....))).....((((.(((((.....))))).))))...........((.((((.((.........))...)))).))...(((((..((....))..))))).. ( -29.20) >consensus GGCAAUCAGUCAAAAACUUUGGCGCAACAUAUGCGUGGGAGGAAGAGAGAUGAUGUCGCACUGAAAAGCAGAGGAAGCCAGGUAAAAGCUGAAGCAGUAGCGCCAGCAAA (((.((((.((.....((((.(((((.....))))).)))).....))..))))...((........)).......)))........((((..((....))..))))... (-27.52 = -27.72 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:19 2006