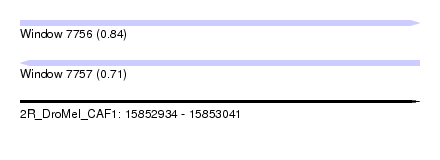
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,852,934 – 15,853,041 |
| Length | 107 |
| Max. P | 0.841000 |

| Location | 15,852,934 – 15,853,041 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15852934 107 + 20766785 -----UUCGUCCAGCUUU--AGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGAGGCAUGCUGCCACGCCCCC- -----..........(((--(((((.(((..((((..((........))..).)))..))).))))))))(((((...((((.((((........)))).)).))...))))).- ( -33.60) >DroSec_CAF1 10342 107 + 1 -----UUUGGCCAGCUCU--AGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGUGGCAUGCUGCCGCGCCCCC- -----((((((.((((((--(((....)))))))))...........((((......))))...))))))(((((...((((.((((((....)))))).)).))...))))).- ( -39.50) >DroEre_CAF1 11734 112 + 1 UUGUAUGUAGUCAGCUUU--AGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGAGGCAUGCUGCCACGCCCCC- ...((((((..(..((((--(((((.(((..((((..((........))..).)))..))).))))))))).)..))))))((((......))))(((.....)))........- ( -34.90) >DroYak_CAF1 11467 107 + 1 -----UGCAGCCCGCUUU--AGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGAGGCAUGCUGCCACGCCCCC- -----.(((((((.((((--(((((.(((..((((..((........))..).)))..))).))))))))).))....((.((((......)))).))..))))).........- ( -39.50) >DroAna_CAF1 11757 113 + 1 CCG--UUUUUCCGGCUCUAUGGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGUGGCAUCCUGUUCCGCCCCCU (((--......))).....((((((.(((..((((..((........))..).)))..))).))))))..(((((.(((.(..((((((....))))))..))))...))))).. ( -37.60) >consensus _____UUUAGCCAGCUUU__AGCUGUUGUUGGAGCUUCGAAUUUGUAUGUUGCCUUUGACACCAGCUAAAGGGGUUACGUGUCUGCCACUUCAGAGGCAUGCUGCCACGCCCCC_ ....................(((((.(((..((((..((........))..).)))..))).)))))...(((((.....(..((((........))))..)......))))).. (-29.44 = -29.28 + -0.16)

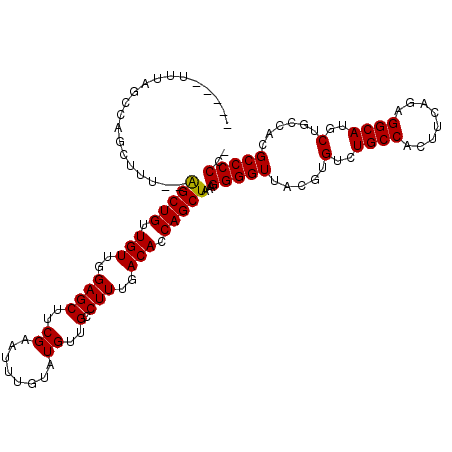
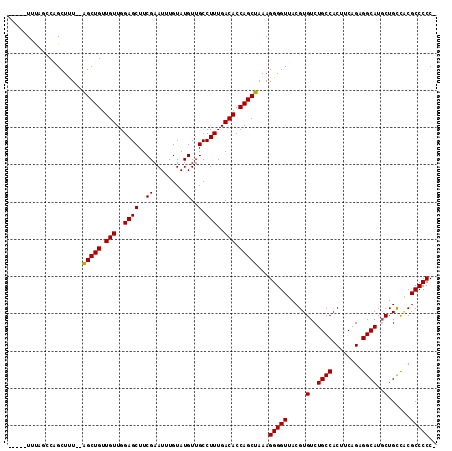
| Location | 15,852,934 – 15,853,041 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15852934 107 - 20766785 -GGGGGCGUGGCAGCAUGCCUCUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCU--AAAGCUGGACGAA----- -(((((((((....)))))))))...(((.....)))....((.(((((((((.(((....(((..(..........)..)))...))).)))))--))))..)).....----- ( -33.00) >DroSec_CAF1 10342 107 - 1 -GGGGGCGCGGCAGCAUGCCACUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCU--AGAGCUGGCCAAA----- -((((..(((......((((((....))))))....)))..))))((((((((.(((....(((..(..........)..)))...))).)))))--)))..........----- ( -35.50) >DroEre_CAF1 11734 112 - 1 -GGGGGCGUGGCAGCAUGCCUCUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCU--AAAGCUGACUACAUACAA -(((((((((....)))))))))...(((((((...........(((((((((.(((....(((..(..........)..)))...))).)))))--))))))).))))...... ( -33.30) >DroYak_CAF1 11467 107 - 1 -GGGGGCGUGGCAGCAUGCCUCUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCU--AAAGCGGGCUGCA----- -(((((((((....)))))))))...(((.....)))(((.((((((((((((.(((....(((..(..........)..)))...))).)))))--)))).))).))).----- ( -40.80) >DroAna_CAF1 11757 113 - 1 AGGGGGCGGAACAGGAUGCCACUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCCAUAGAGCCGGAAAAA--CGG ((((((((.....(..((((((....))))))..).)))..)))))...((((.(((....(((..(..........)..)))...))).)))).......(((......--))) ( -31.20) >consensus _GGGGGCGUGGCAGCAUGCCUCUGAAGUGGCAGACACGUAACCCCUUUAGCUGGUGUCAAAGGCAACAUACAAAUUCGAAGCUCCAACAACAGCU__AAAGCUGGCCAAA_____ ..((((((.(((.....))).)(((((((.....)))(((...(((((..(....)..))))).....)))...))))..))))).....((((......))))........... (-25.64 = -26.44 + 0.80)

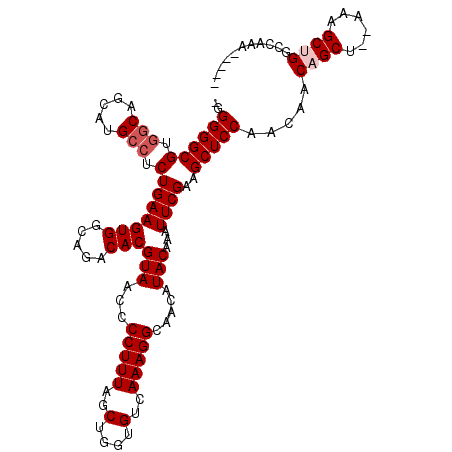
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:15 2006