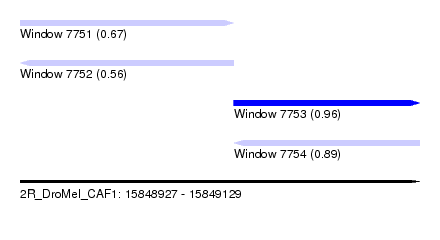
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,848,927 – 15,849,129 |
| Length | 202 |
| Max. P | 0.963058 |

| Location | 15,848,927 – 15,849,035 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -12.68 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15848927 108 + 20766785 CUUCCCCUCCACCCCCCGC----CAAUUUUCC--GCCAACUCUCAACACAUUUUGGCCUGCCCCACCCACUUU-UCUUAGUCCGGGGGCGUUGGCAGAUAACGGUUUACUUUCUU .................((----(.....((.--((((((..((((......))))...(((((....(((..-....)))...))))))))))).))....))).......... ( -25.40) >DroSec_CAF1 6146 101 + 1 CUCCCCCUGCA-------A----AAAUUUUUU--ACCAACUCUCACCACAUUUUGGCCGGCCCCACCCACUUU-UCUUAGCCCGGGGGCGUUGGCAGAUAACGGUUUACUUUCUU ...........-------.----.........--..........(((....((((.((((((((.((..((..-....))...))))).)))))))))....))).......... ( -18.50) >DroSim_CAF1 6065 96 + 1 CUCCCCCUGCA-------A----CAAUUUUC-------ACUCUUACCACAUUUUGGCCUGCCCCACCCACUUU-UCUUAGCCCGGGGGCGUUGGCAGAUAACGGUUUACUUUCUU ......(((((-------(----(.......-------.......(((.....)))...(((((.....((..-....))....)))))))).)))).................. ( -19.40) >DroEre_CAF1 7506 103 + 1 CCUCCCCCCCA-------C----CGACCUUCCGACCCCCCUCGCGCCACAUUUUGGAUUGCCCCACCCACUUCUUCUUAGCCCGGGGGCGUUGGCA-AUAACGGUUUACUUUCUU ..........(-------(----((.....((((((((((..((.........(((..........)))..........))..))))).)))))..-....)))).......... ( -24.61) >DroYak_CAF1 7186 103 + 1 CCGCCCCCUCA-------GUUUUCCGCCUUC---CCCCCCUCACAUCACAUUCUGGAUUGCCCCGCCCACUUC-UCUUAGCCCGGGGGCGUUGGCA-AUAACGGUUUACUUUCUU ..........(-------((...(((.....---....((..............))((((((.(((((.((..-.........)))))))..))))-))..)))...)))..... ( -21.44) >consensus CUCCCCCUCCA_______A____CAAUUUUC___CCCAACUCUCACCACAUUUUGGCCUGCCCCACCCACUUU_UCUUAGCCCGGGGGCGUUGGCAGAUAACGGUUUACUUUCUU ............................................(((............(((((.....((.......))....)))))((((.....))))))).......... (-12.68 = -12.60 + -0.08)

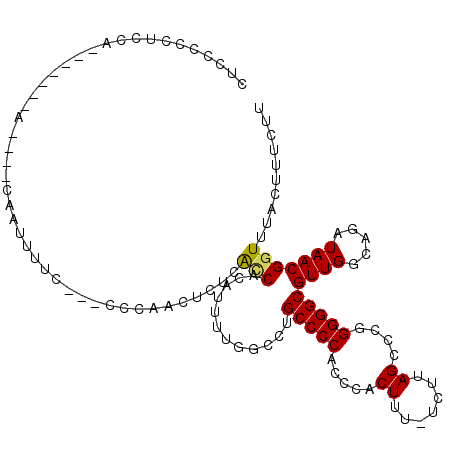
| Location | 15,848,927 – 15,849,035 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15848927 108 - 20766785 AAGAAAGUAAACCGUUAUCUGCCAACGCCCCCGGACUAAGA-AAAGUGGGUGGGGCAGGCCAAAAUGUGUUGAGAGUUGGC--GGAAAAUUG----GCGGGGGGUGGAGGGGAAG ...........(((((((((((((((((((((..(((....-..)))..).)))))....(((......)))...))))))--)))....))----))))............... ( -34.40) >DroSec_CAF1 6146 101 - 1 AAGAAAGUAAACCGUUAUCUGCCAACGCCCCCGGGCUAAGA-AAAGUGGGUGGGGCCGGCCAAAAUGUGGUGAGAGUUGGU--AAAAAAUUU----U-------UGCAGGGGGAG ......(((((..(((.(.(((((((((((((..(((....-..)))..).)))))..((((.....))))....))))))--).).))).)----)-------)))........ ( -27.40) >DroSim_CAF1 6065 96 - 1 AAGAAAGUAAACCGUUAUCUGCCAACGCCCCCGGGCUAAGA-AAAGUGGGUGGGGCAGGCCAAAAUGUGGUAAGAGU-------GAAAAUUG----U-------UGCAGGGGGAG ...........((....(((((.(((((((((..(((....-..)))..).)))))..((((.....))))......-------.......)----)-------)))))).)).. ( -26.20) >DroEre_CAF1 7506 103 - 1 AAGAAAGUAAACCGUUAU-UGCCAACGCCCCCGGGCUAAGAAGAAGUGGGUGGGGCAAUCCAAAAUGUGGCGCGAGGGGGGUCGGAAGGUCG----G-------UGGGGGGGAGG ...........((.((.(-..((....((((((.((((........(((((......))))).....)))).)).))))(..(....)..))----)-------..))).))... ( -35.42) >DroYak_CAF1 7186 103 - 1 AAGAAAGUAAACCGUUAU-UGCCAACGCCCCCGGGCUAAGA-GAAGUGGGCGGGGCAAUCCAGAAUGUGAUGUGAGGGGGG---GAAGGCGGAAAAC-------UGAGGGGGCGG ......((...((.((((-((((..(.(((((..(((....-......)))(((....)))..............))))).---)..))))).....-------))).)).)).. ( -28.50) >consensus AAGAAAGUAAACCGUUAUCUGCCAACGCCCCCGGGCUAAGA_AAAGUGGGUGGGGCAGGCCAAAAUGUGGUGAGAGUGGGG___GAAAAUUG____G_______UGCAGGGGAAG ...........((.((..(((((..(((((....(((.......)))))))).)))))((((.....))))....................................)).))... (-15.50 = -16.34 + 0.84)

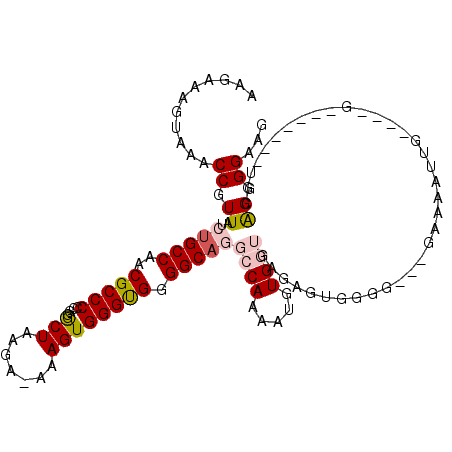
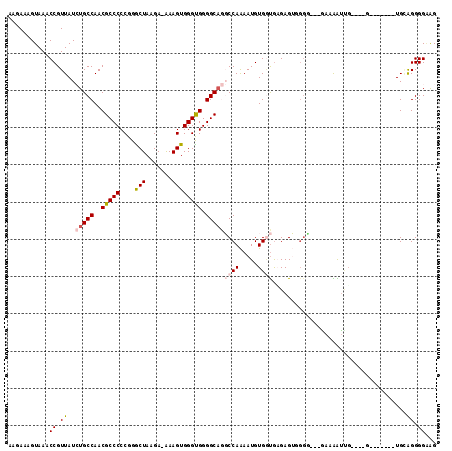
| Location | 15,849,035 – 15,849,129 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -11.27 |
| Energy contribution | -12.34 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15849035 94 + 20766785 AAUCUCCGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGUAUGUA----UGGCGCAUUCUUUCCGCUUCUGCGAAAUAACAUACGAUCUUCU-------UCAU .(((.(((((((((....(((((((.....))..)))))..))))))----)))...........(((....)))...........))).....-------.... ( -20.00) >DroVir_CAF1 9809 78 + 1 AAUCUUUGUACAUAAUUUUUGCAACCUCAUGUUACGCAA--------UUUCGGGG---UUGUUGCCGCAUCCGUGUG---------GCUGUACU-------CUAU .......(((((........((((((((..(((....))--------)...))))---)))).((((((....))))---------))))))).-------.... ( -22.60) >DroSec_CAF1 6247 94 + 1 AAUCUCUGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGUAUGUA----UGGCGCAUUCUUUCCGCUUCUGCGGAAUAACACACGCUCUUCU-------CCAU .......(((((((....(((((((.....))..)))))..))))))----)((((......((((((....)))))).......)))).....-------.... ( -24.52) >DroSim_CAF1 6161 94 + 1 AAUCUCCGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGUAUGUA----UGGCGCAUUCUUUCCGCUUCUGCGGAAUAACACACGCUCUUCU-------CCAU ......((((((((....(((((((.....))..)))))..))))))----))(((......((((((....)))))).......)))......-------.... ( -26.42) >DroEre_CAF1 7609 97 + 1 AAUCUCUGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGU----A----UGGCGCAUUCUUUCCGCUUCUGCGGAAUAACAUACGCUCUUCUUUAGCUGCCAU .......((((((......((((((.....))..))))))))----)----)(((.......((((((....))))))........(((.......))).))).. ( -22.00) >DroYak_CAF1 7289 97 + 1 AAUCUCCGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGU----A----UGGCGCAUUCUUUCCGCUUCUGCGGAAUAAAAUACGCUUAUCUUCAGCUGCCAU .....((((((((......((((((.....))..))))))))----)----))).(((....((((((....))))))........(((.......))))))... ( -23.60) >consensus AAUCUCCGUACAUAAUUUUUGCGACUUCAUGUUACGCAAUGU____A____UGGCGCAUUCUUUCCGCUUCUGCGGAAUAACAUACGCUCUUCU_______CCAU ..................(((((((.....))..))))).............((((......((((((....)))))).......))))................ (-11.27 = -12.34 + 1.07)
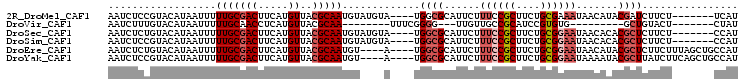
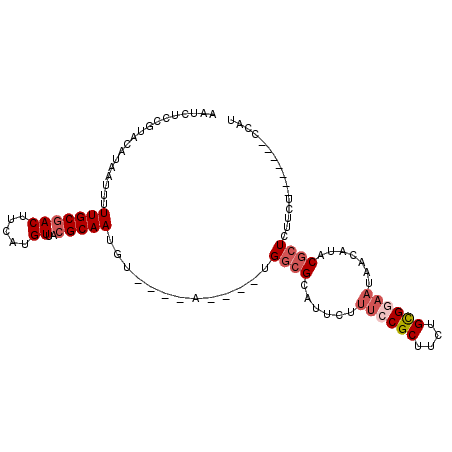
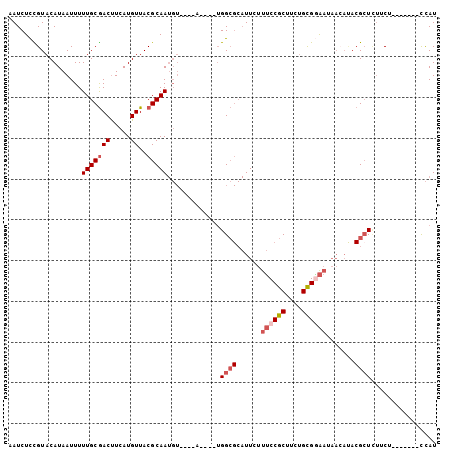
| Location | 15,849,035 – 15,849,129 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15849035 94 - 20766785 AUGA-------AGAAGAUCGUAUGUUAUUUCGCAGAAGCGGAAAGAAUGCGCCA----UACAUACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACGGAGAUU ....-------....((((((((....((((((....))))))...)))).((.----((((((..(((((...........)))))....)))))).)).)))) ( -21.70) >DroVir_CAF1 9809 78 - 1 AUAG-------AGUACAGC---------CACACGGAUGCGGCAACAA---CCCCGAAA--------UUGCGUAACAUGAGGUUGCAAAAAUUAUGUACAAAGAUU ....-------.(((((((---------(.((....)).))).....---........--------....(((((.....)))))........)))))....... ( -15.70) >DroSec_CAF1 6247 94 - 1 AUGG-------AGAAGAGCGUGUGUUAUUCCGCAGAAGCGGAAAGAAUGCGCCA----UACAUACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACAGAGAUU ....-------....(.((((((....((((((....))))))...))))))).----((((((..(((((...........)))))....))))))........ ( -23.70) >DroSim_CAF1 6161 94 - 1 AUGG-------AGAAGAGCGUGUGUUAUUCCGCAGAAGCGGAAAGAAUGCGCCA----UACAUACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACGGAGAUU ....-------....(.((((((....((((((....))))))...))))))).----((((((..(((((...........)))))....))))))........ ( -23.70) >DroEre_CAF1 7609 97 - 1 AUGGCAGCUAAAGAAGAGCGUAUGUUAUUCCGCAGAAGCGGAAAGAAUGCGCCA----U----ACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACAGAGAUU (((((.(((.......)))((((....((((((....))))))...))))))))----)----...(((((...........))))).................. ( -25.80) >DroYak_CAF1 7289 97 - 1 AUGGCAGCUGAAGAUAAGCGUAUUUUAUUCCGCAGAAGCGGAAAGAAUGCGCCA----U----ACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACGGAGAUU (((((.(((.......)))(((((((.((((((....)))))))))))))))))----)----...((.((((.(((((...........))))))))).))... ( -30.90) >consensus AUGG_______AGAAGAGCGUAUGUUAUUCCGCAGAAGCGGAAAGAAUGCGCCA____U____ACAUUGCGUAACAUGAAGUCGCAAAAAUUAUGUACAGAGAUU .................((((((....((((((....))))))...))))))..............(((((...........))))).................. (-13.91 = -14.78 + 0.88)

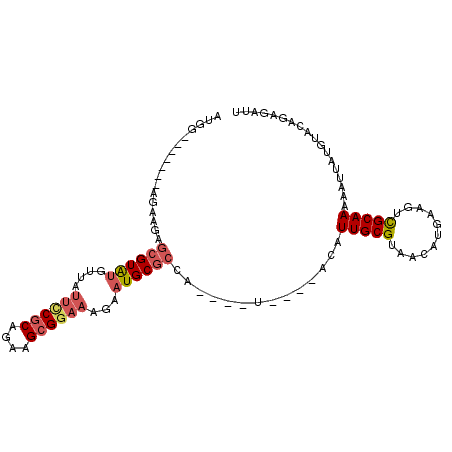
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:12 2006