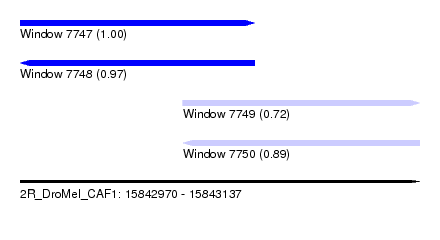
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,842,970 – 15,843,137 |
| Length | 167 |
| Max. P | 0.996748 |

| Location | 15,842,970 – 15,843,068 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.06 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15842970 98 + 20766785 GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCAAGCAACUGCAUAGAAGUGGGAAAACCCAAUUACCUCG ((((((.....))))))........(((((...........(((((((..........(((......))).)))))))(((....)))...))))).. ( -24.50) >DroSec_CAF1 56 98 + 1 GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCAAGCAACUGCAUAGAAGUGGGAAAACCCAAAUGCCUCG ((((((.....)))))).((((..(((....))).......(((((((..........(((......))).)))))))(((....)))..)))).... ( -26.40) >DroSim_CAF1 51 98 + 1 GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCAAGCAACUGUAUAGAAGUGGGAAAACCCAAAUGCCUCG ((((((.....)))))).((((..(((....))).......(((((((.....(((.((.......))))))))))))(((....)))..)))).... ( -26.20) >DroEre_CAF1 1345 96 + 1 GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCA-GCAACUGCAUAGAAGUGGGAAAACCCA-AUGCCUCG ((((((.....)))))).((((..(((....))).......(((((((..........(((-(...)))).)))))))(((....))).-)))).... ( -27.70) >DroYak_CAF1 51 97 + 1 GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCA-GCAACUGCAUAGAAGUGGGAAAACCCAAAUGCCUCG ((((((.....)))))).((((..(((....))).......(((((((..........(((-(...)))).)))))))(((....)))..)))).... ( -27.70) >consensus GGCAGCCGACAGCUGCCCGCAUAACAGGUACCUGAACUGAAACUUCUACCAACUACCAGCAAGCAACUGCAUAGAAGUGGGAAAACCCAAAUGCCUCG ((((((.....))))))........(((((...........(((((((..........(((......))).)))))))(((....)))...))))).. (-24.38 = -24.06 + -0.32)


| Location | 15,842,970 – 15,843,068 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -33.22 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15842970 98 - 20766785 CGAGGUAAUUGGGUUUUCCCACUUCUAUGCAGUUGCUUGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC ..(((((.(((..((.....((((((((.((..((((....))))..))))))))))...))..))).)))))........((((((.....)))))) ( -33.10) >DroSec_CAF1 56 98 - 1 CGAGGCAUUUGGGUUUUCCCACUUCUAUGCAGUUGCUUGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC ...((((..((((....)))).......((((((((((((.((((.((((..((....))...)))).))))......))))))))))))....)))) ( -34.80) >DroSim_CAF1 51 98 - 1 CGAGGCAUUUGGGUUUUCCCACUUCUAUACAGUUGCUUGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC ...((((..((((....)))).......((((((((((((.((((.((((..((....))...)))).))))......))))))))))))....)))) ( -34.50) >DroEre_CAF1 1345 96 - 1 CGAGGCAU-UGGGUUUUCCCACUUCUAUGCAGUUGC-UGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC .(((((..-((((....)))).((((((.((..(((-.....)))..))))))))))))).((.(((....)))....)).((((((.....)))))) ( -32.00) >DroYak_CAF1 51 97 - 1 CGAGGCAUUUGGGUUUUCCCACUUCUAUGCAGUUGC-UGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC .(((((...((((....)))).((((((.((..(((-.....)))..))))))))))))).((.(((....)))....)).((((((.....)))))) ( -32.00) >consensus CGAGGCAUUUGGGUUUUCCCACUUCUAUGCAGUUGCUUGCUGGUAGUUGGUAGAAGUUUCAGUUCAGGUACCUGUUAUGCGGGCAGCUGUCGGCUGCC ...((((..((((....)))).......((((((((((((.((((.((((..((....))...)))).))))......))))))))))))....)))) (-33.22 = -33.30 + 0.08)

| Location | 15,843,038 – 15,843,137 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
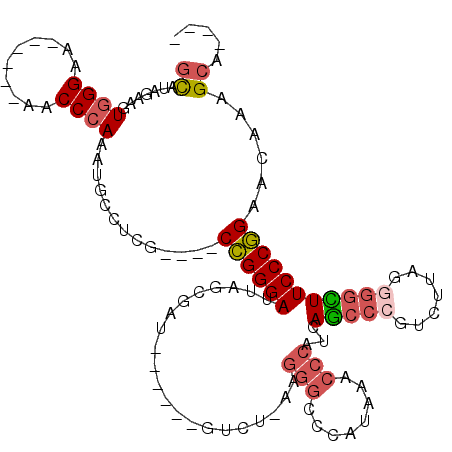
>2R_DroMel_CAF1 15843038 99 + 20766785 GCAUAGAAGUGGGAA------AACCCAAUUACCUCG----CCGGGAUUAGCGAU------GUCU-AAGGGCCCAUAAACCCAUCAGCCCGUCUUAGGGGCUUCCCGGAACAAAGCA---- ((.......((((..------..)))).........----((((((..(((...------....-..(((........))).....(((......))))))))))))......)).---- ( -30.80) >DroSec_CAF1 124 99 + 1 GCAUAGAAGUGGGAA------AACCCAAAUGCCUCG----CCGGGAUUAGCGAU------GUCU-AAGGGCCCAUAAACCCAUCAGCCCGUCUUAGGGGCUUCCCGGAACAAAGCA---- ((((.....((((..------..)))).))))....----((((((..(((...------....-..(((........))).....(((......)))))))))))).........---- ( -31.00) >DroSim_CAF1 119 99 + 1 GUAUAGAAGUGGGAA------AACCCAAAUGCCUCG----CCGGGAUUAGCGAU------GUCU-AAGGGCCCAUAAACCCAUCAGCCCGUCUUAGGGGCUUCCCGGAACAAAGCA---- .........((((..------..))))..(((....----((((((..(((...------....-..(((........))).....(((......))))))))))))......)))---- ( -30.00) >DroEre_CAF1 1412 98 + 1 GCAUAGAAGUGGGAA------AACCCA-AUGCCUCG----CCGGGAUUAGCGAU------GUCU-AAGGGCCCAUAAACCCAUCAGCCCGCCUUAGGGGCUUCCCGGAACCAAGCA---- ((.......((((..------..))))-........----((((((..(((...------....-..(((........))).....(((......))))))))))))......)).---- ( -30.80) >DroYak_CAF1 118 99 + 1 GCAUAGAAGUGGGAA------AACCCAAAUGCCUCG----CCGGGAUUAGCGAU------GUCU-AAGGGCCCAUAAACCCAUCAGCCGGUCUUAAGGGUUUCCCGGAACUAAGCA---- ((.(((...((((..------..)))).........----((((((((((....------..))-)).(((((.(((..((.......))..))).)))))))))))..))).)).---- ( -29.70) >DroAna_CAF1 111 118 + 1 GCCUAGAACUAGGAAUCCUCCAACCAAA--ACCUCGCUGGCUGGGAUUAGCCAAAAGCCGGCCGGACGGACCCCUAAUAAAAUAAUAAAAACUAAAGGGAUUCCCAGGAAGGCGGAUCAG .((((....)))).((((.((..((...--.((..(((((((.((.....))...))))))).))..(((.((((....................))))..)))..))..)).))))... ( -35.95) >consensus GCAUAGAAGUGGGAA______AACCCAAAUGCCUCG____CCGGGAUUAGCGAU______GUCU_AAGGGCCCAUAAACCCAUCAGCCCGUCUUAGGGGCUUCCCGGAACAAAGCA____ ((.......((((..........)))).............((((((.....................(((........)))...(((((.......)))))))))))......))..... (-21.02 = -21.72 + 0.70)

| Location | 15,843,038 – 15,843,137 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -23.88 |
| Energy contribution | -26.02 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
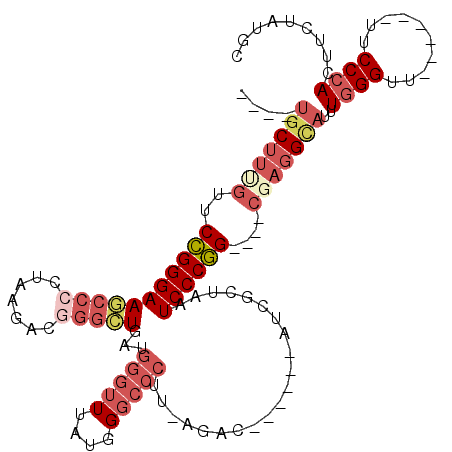
>2R_DroMel_CAF1 15843038 99 - 20766785 ----UGCUUUGUUCCGGGAAGCCCCUAAGACGGGCUGAUGGGUUUAUGGGCCCUU-AGAC------AUCGCUAAUCCCGG----CGAGGUAAUUGGGUU------UUCCCACUUCUAUGC ----(((((((..((((((((((((......)))((((.(((((....)))))))-))..------...)))..))))))----)))))))..((((..------..))))......... ( -36.00) >DroSec_CAF1 124 99 - 1 ----UGCUUUGUUCCGGGAAGCCCCUAAGACGGGCUGAUGGGUUUAUGGGCCCUU-AGAC------AUCGCUAAUCCCGG----CGAGGCAUUUGGGUU------UUCCCACUUCUAUGC ----(((((((..((((((((((((......)))((((.(((((....)))))))-))..------...)))..))))))----)))))))..((((..------..))))......... ( -37.80) >DroSim_CAF1 119 99 - 1 ----UGCUUUGUUCCGGGAAGCCCCUAAGACGGGCUGAUGGGUUUAUGGGCCCUU-AGAC------AUCGCUAAUCCCGG----CGAGGCAUUUGGGUU------UUCCCACUUCUAUAC ----(((((((..((((((((((((......)))((((.(((((....)))))))-))..------...)))..))))))----)))))))..((((..------..))))......... ( -37.80) >DroEre_CAF1 1412 98 - 1 ----UGCUUGGUUCCGGGAAGCCCCUAAGGCGGGCUGAUGGGUUUAUGGGCCCUU-AGAC------AUCGCUAAUCCCGG----CGAGGCAU-UGGGUU------UUCCCACUUCUAUGC ----(((((.(..((((((.(((.....)))(((((.(((....))).)))))..-....------........))))))----).))))).-((((..------..))))......... ( -36.50) >DroYak_CAF1 118 99 - 1 ----UGCUUAGUUCCGGGAAACCCUUAAGACCGGCUGAUGGGUUUAUGGGCCCUU-AGAC------AUCGCUAAUCCCGG----CGAGGCAUUUGGGUU------UUCCCACUUCUAUGC ----.((.(((....(((((((((..((..(((.((((.(((((....)))))))-)).)------.(((((......))----)))))..)).)))))------)))).....))).)) ( -33.80) >DroAna_CAF1 111 118 - 1 CUGAUCCGCCUUCCUGGGAAUCCCUUUAGUUUUUAUUAUUUUAUUAGGGGUCCGUCCGGCCGGCUUUUGGCUAAUCCCAGCCAGCGAGGU--UUUGGUUGGAGGAUUCCUAGUUCUAGGC .......((((..(((((((((((((((((............))))))).....((((((((((((((((((......)))))..)))))--..))))))))))))))))))....)))) ( -43.30) >consensus ____UGCUUUGUUCCGGGAAGCCCCUAAGACGGGCUGAUGGGUUUAUGGGCCCUU_AGAC______AUCGCUAAUCCCGG____CGAGGCAUUUGGGUU______UUCCCACUUCUAUGC ....(((((((..(((((((((((.......)))))...(((((....))))).....................))))))....)))))))..((((..........))))......... (-23.88 = -26.02 + 2.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:08 2006