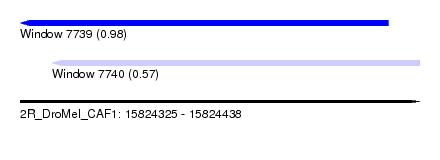
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,824,325 – 15,824,438 |
| Length | 113 |
| Max. P | 0.983680 |

| Location | 15,824,325 – 15,824,429 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
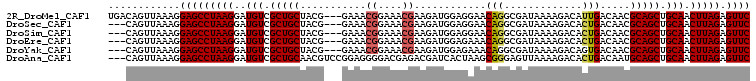
>2R_DroMel_CAF1 15824325 104 - 20766785 AAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACAUUGACAACGCAGCUGCAACUUAGAGUUCCGAACCUAU ..((((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))))........ ( -31.80) >DroSec_CAF1 13541 104 - 1 AAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAACCGUU ..((((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))))........ ( -31.80) >DroSim_CAF1 16950 104 - 1 AAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAACCGUU ..((((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))))........ ( -31.80) >DroEre_CAF1 9288 104 - 1 AAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAACCGUU ..((((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))))........ ( -31.80) >DroYak_CAF1 16840 104 - 1 AAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACAGUGACAACGCAGCUGCAACUUAGAGUUCCGAACCGUU ..((((((((((..((((((((((.((---....))....((....((......))..)).....)).))))))))..((....))..))))).)))))........ ( -34.00) >DroAna_CAF1 4531 107 - 1 AAGGAGCCUAAGGAUGUCGCUGCAACGUCCGGAGGGGACGAGACGAUCACUAAGCGGGAGUUAAAAGACACUGACAAUGCAGCUGCAACUUAGAGUUCCGAAUUUUU ..((((((((((..(((.((((((.(((((.....))))).............((((..(((....))).))).)..)))))).))).))))).)))))........ ( -37.90) >consensus AAGGAGCCUAAGGAUGUCGCUGCUACG___GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAACCGUU ..((((((((((..(((.(((((...........((....))............(((.............))).....))))).))).))))).)))))........ (-28.14 = -28.39 + 0.25)

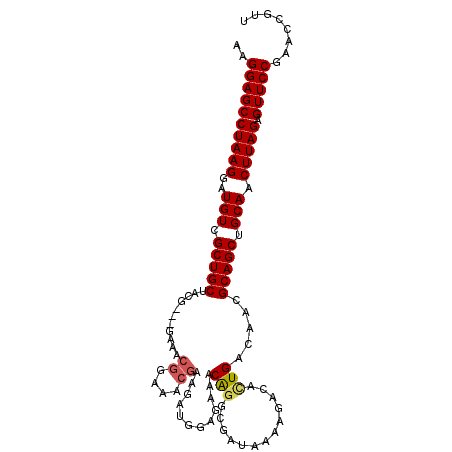
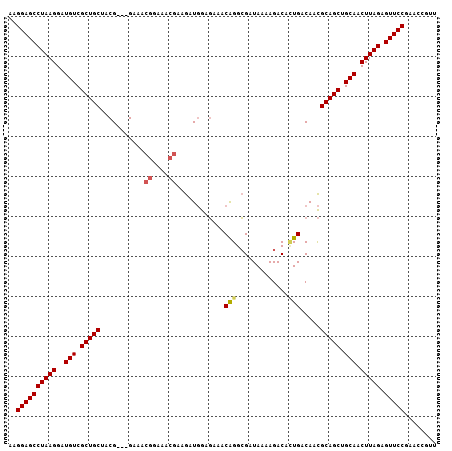
| Location | 15,824,334 – 15,824,438 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -22.93 |
| Energy contribution | -23.19 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15824334 104 - 20766785 UGACAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACAUUGACAACGCAGCUGCAACUUAGAGUUC ............(((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))) ( -26.60) >DroSec_CAF1 13550 101 - 1 ---CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUC ---.........(((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))) ( -26.60) >DroSim_CAF1 16959 101 - 1 ---CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUC ---.........(((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))) ( -26.60) >DroEre_CAF1 9297 101 - 1 ---CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUC ---.........(((((((((..(((.(((((..((---....))....((....((......))..))..................))))).))).))))).)))) ( -26.60) >DroYak_CAF1 16849 101 - 1 ---CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACAGUGACAACGCAGCUGCAACUUAGAGUUC ---.........(((((((((..((((((((((.((---....))....((....((......))..)).....)).))))))))..((....))..))))).)))) ( -28.80) >DroAna_CAF1 4540 104 - 1 ---CAGUUAAAGGAGCCUAAGGAUGUCGCUGCAACGUCCGGAGGGGACGAGACGAUCACUAAGCGGGAGUUAAAAGACACUGACAAUGCAGCUGCAACUUAGAGUUC ---.........(((((((((..(((.((((((.(((((.....))))).............((((..(((....))).))).)..)))))).))).))))).)))) ( -32.70) >consensus ___CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG___GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAAAAGACACUGACAACGCAGCUGCAACUUAGAGUUC ............(((((((((..(((.(((((...........((....))............(((.............))).....))))).))).))))).)))) (-22.93 = -23.19 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:59 2006