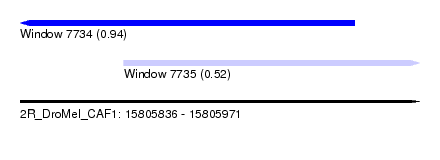
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,805,836 – 15,805,971 |
| Length | 135 |
| Max. P | 0.935271 |

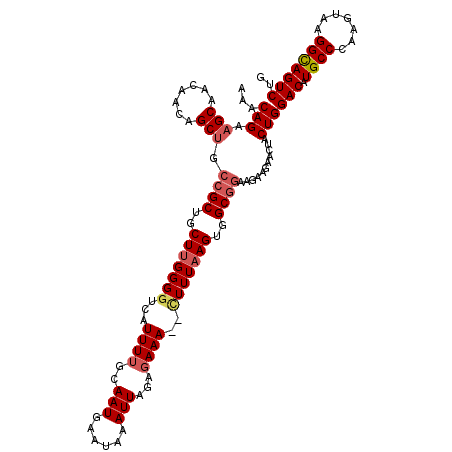
| Location | 15,805,836 – 15,805,949 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -26.35 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15805836 113 - 20766785 AAACAGAAGCAACAACAGCUGCCGCUGCUUGGGGUCAUUUUGCAAUGAAUAAAUUAGAGAAA--CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...(((.(((.......))).((((..((((((((..((((..(((......))).)))).)--)))))))..))))...........)))(((.((((.......))))))).. ( -32.20) >DroSec_CAF1 3845 113 - 1 AAACAGAUGCAACAACAGCUGCCGCUGCUUGGGGUCAUUUUGCAAUGAAUAAAUUAGAGAAA--CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...............((((((((..((((((((.(((((....))))).....((((((...--))))))(((.(((.((.....)).)))..))).)))))))).))))).))) ( -31.30) >DroSim_CAF1 3230 113 - 1 AAACAGAAGCAACAACAGCUGCCGCUGCUUGGGGUCAUUUUGCAAUGAAUAAAUUAGAGAAA--CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...(((.(((.......))).((((..((((((((..((((..(((......))).)))).)--)))))))..))))...........)))(((.((((.......))))))).. ( -32.20) >DroEre_CAF1 4163 113 - 1 AAACAGAAGCAACAACAGCUGCCGCUGCUUGGGGUCAUUUUGCAAUGAAUAAAUUAGAGAAA--CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...(((.(((.......))).((((..((((((((..((((..(((......))).)))).)--)))))))..))))...........)))(((.((((.......))))))).. ( -32.20) >DroYak_CAF1 3255 112 - 1 AAACAGAAGCAACAACAGCUGCCGCUGCU-GGGAUCAUUUUGCAAUGAAUAAAUUAGAGAAA--CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...(((.(((.......))).((((..((-....(((((....)))))......(((((...--)))))))..))))...........)))(((.((((.......))))))).. ( -27.10) >DroAna_CAF1 3819 110 - 1 AAACAGAAGCAGA---AGCUG--GCUGCUUGGGGUCAUUUUGCAAUGCAUAAAUUAGACAAAAGUUUUAAGUGGCGGAAGAAGAACAACUGGACAUGCCCAAGUACGGUAGUCGA ...(((.(((...---.))).--((..(((((((.(.(((((((((......))).).)))))))))))))..)).............)))(((.((((.......))))))).. ( -24.00) >consensus AAACAGAAGCAACAACAGCUGCCGCUGCUUGGGGUCAUUUUGCAAUGAAUAAAUUAGAGAAA__CUUUAAGUGGCGGAAGAAGAACUACUGGACAUGCCCAAGUAAGGCAGUCUG ...(((.(((.......))).((((..(((((((...((((..(((......)))...))))..)))))))..))))...........)))(((.((((.......))))))).. (-26.35 = -27.10 + 0.75)

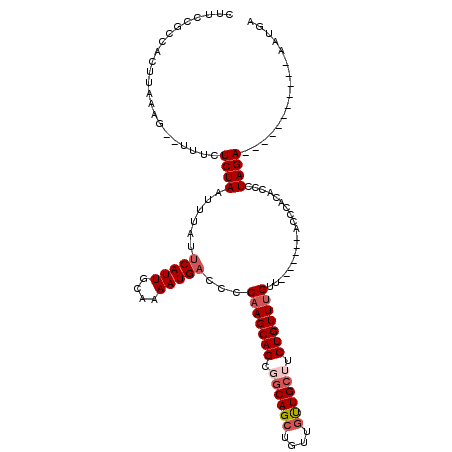
| Location | 15,805,871 – 15,805,971 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15805871 100 + 20766785 CUUCCGCCACUUAAAG--UUUCUCUAAUUUAUUCAUUGCAAAAUGACCCCAAGCAGCGGCAGCUGUUGUUGCUUCUGUUUGUU-------ACCCACACCCUAGA-----------AAUGA ...............(--(((((.........(((((....)))))...(((((((.((((((....)))))).)))))))..-------...........)))-----------))).. ( -23.10) >DroSec_CAF1 3880 100 + 1 CUUCCGCCACUUAAAG--UUUCUCUAAUUUAUUCAUUGCAAAAUGACCCCAAGCAGCGGCAGCUGUUGUUGCAUCUGUUUGUU-------ACCCACACCCUAGA-----------AAUGA ...............(--(((((.........(((((....)))))...(((((((..(((((....)))))..)))))))..-------...........)))-----------))).. ( -20.90) >DroSim_CAF1 3265 100 + 1 CUUCCGCCACUUAAAG--UUUCUCUAAUUUAUUCAUUGCAAAAUGACCCCAAGCAGCGGCAGCUGUUGUUGCUUCUGUUUGUU-------ACCCACACCCUAGA-----------AAUGA ...............(--(((((.........(((((....)))))...(((((((.((((((....)))))).)))))))..-------...........)))-----------))).. ( -23.10) >DroEre_CAF1 4198 100 + 1 CUUCCGCCACUUAAAG--UUUCUCUAAUUUAUUCAUUGCAAAAUGACCCCAAGCAGCGGCAGCUGUUGUUGCUUCUGUUUGUU-------ACCCACACCCUAGA-----------AAUGU ...............(--(((((.........(((((....)))))...(((((((.((((((....)))))).)))))))..-------...........)))-----------))).. ( -23.10) >DroYak_CAF1 3290 101 + 1 CUUCCGCCACUUAAAG--UUUCUCUAAUUUAUUCAUUGCAAAAUGAUCCC-AGCAGCGGCAGCUGUUGUUGCUUCUGUUUGUU-----UUUCCCACACCCUAGA-----------AAUGU ...............(--(((((.........(((((....)))))....-(((((.((((((....)))))).)))))....-----.............)))-----------))).. ( -19.20) >DroAna_CAF1 3854 115 + 1 CUUCCGCCACUUAAAACUUUUGUCUAAUUUAUGCAUUGCAAAAUGACCCCAAGCAGC--CAGCU---UCUGCUUCUGUUUGUUCGCAGUUACCCACUCCCUAGAACGACCAUUAAAAAGU ...............(((((((((.........((((....))))....(((((((.--.(((.---...))).))))))).........................))).....)))))) ( -15.10) >consensus CUUCCGCCACUUAAAG__UUUCUCUAAUUUAUUCAUUGCAAAAUGACCCCAAGCAGCGGCAGCUGUUGUUGCUUCUGUUUGUU_______ACCCACACCCUAGA___________AAUGA ......................((((......(((((....)))))...(((((((.((((((....)))))).)))))))...................))))................ (-17.42 = -18.28 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:55 2006