| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,794,138 – 15,794,230 |
| Length | 92 |
| Max. P | 0.562519 |

| Location | 15,794,138 – 15,794,230 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.37 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15794138 92 + 20766785 CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAUUGGA-AAAAAGCGAAUGU-UUUAGAUGAG--- .((((.((((((....(((((((((.....))))...)))))...)))))))).)).(((......)))-..(((((....))-))).......--- ( -29.40) >DroSec_CAF1 109 95 + 1 CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAAUAAAAACAAAGCGAAAGU-UUUAAUUGAGAU- .((((.((((((....(((((((((.....))))...)))))...)))))))).))................(((((....))-))).........- ( -30.60) >DroSim_CAF1 109 93 + 1 CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAAUCAAAAAAAAGCGACCGU-UUUAGAAGAG--- .((((.((((((....(((((((((.....))))...)))))...)))))))).))...(((((((...............))-))))).....--- ( -27.76) >DroEre_CAF1 148 95 + 1 CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAAUAGGAAAAACGUGAUAGU-UUCAGAUGCUGG- (((((.((((((....(((((((((.....))))...)))))...))))))))....(((((...))))).((((......))-))))).......- ( -30.90) >DroYak_CAF1 109 96 + 1 CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAAUGUAAAAAAGUCGUCAGUUUUCAGUUGCUGG- .((((.((((((....(((((((((.....))))...)))))...)))))))).)).......................((((........)))).- ( -28.90) >DroPer_CAF1 109 87 + 1 CUGUGCAGCAGUCAGAGUACCGAUCAUCACAAUCCUAGGCACCAAUCCGCCCCAGAGUCCUGUAGAAA-----AGU--AGAGU-UUAAGGCAAG--G .(.(((((.(.((.((.......))............(((........)))...)).).))))).)..-----...--.....-..........--. ( -10.60) >consensus CUGGGCGGCGGUCAAUGUGCCGAUCAUCACGAUCCUGGGCACCAAACCGCCCCACAAUCCUGAAAUAAAAAAAAAGCGAAAGU_UUUAGAUGAG___ ...((.((((((....(((((((((.....))))...)))))...))))))))............................................ (-19.37 = -20.37 + 1.00)

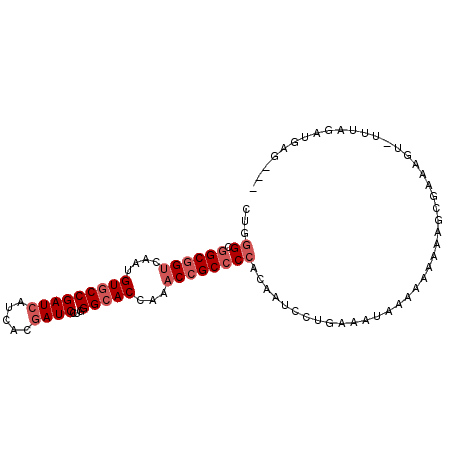
| Location | 15,794,138 – 15,794,230 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.48 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15794138 92 - 20766785 ---CUCAUCUAAA-ACAUUCGCUUUUU-UCCAAUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG ---..........-.............-(((......))).((.(((((((((..(((((...((((.....)))))))))...))))))).)))). ( -30.90) >DroSec_CAF1 109 95 - 1 -AUCUCAAUUAAA-ACUUUCGCUUUGUUUUUAUUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG -((((.(((.(((-((.........))))).)))...))))((.(((((((((..(((((...((((.....)))))))))...))))))).)))). ( -30.60) >DroSim_CAF1 109 93 - 1 ---CUCUUCUAAA-ACGGUCGCUUUUUUUUGAUUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG ---..........-..((((((....(((((....)))))..))).(((((((..(((((...((((.....)))))))))...))))))))))... ( -31.80) >DroEre_CAF1 148 95 - 1 -CCAGCAUCUGAA-ACUAUCACGUUUUUCCUAUUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG -...((.......-....(((((....((((.....)))).)))))(((((((..(((((...((((.....)))))))))...))))))))).... ( -34.40) >DroYak_CAF1 109 96 - 1 -CCAGCAACUGAAAACUGACGACUUUUUUACAUUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG -.......((((((..(((........)))..))))))...((.(((((((((..(((((...((((.....)))))))))...))))))).)))). ( -32.80) >DroPer_CAF1 109 87 - 1 C--CUUGCCUUAA-ACUCU--ACU-----UUUCUACAGGACUCUGGGGCGGAUUGGUGCCUAGGAUUGUGAUGAUCGGUACUCUGACUGCUGCACAG .--((((....((-(....--..)-----))....)))).((.((.(((((...((((((...((((.....))))))))))....))))).)).)) ( -17.90) >consensus ___CUCAUCUAAA_ACUGUCGCUUUUUUUUUAUUUCAGGAUUGUGGGGCGGUUUGGUGCCCAGGAUCGUGAUGAUCGGCACAUUGACCGCCGCCCAG ........................................(((.(.(((((((..(((((...((((.....)))))))))...))))))).).))) (-25.84 = -25.48 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:50 2006