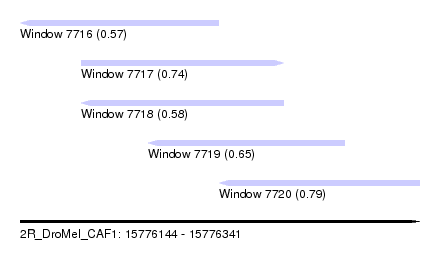
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,776,144 – 15,776,341 |
| Length | 197 |
| Max. P | 0.794465 |

| Location | 15,776,144 – 15,776,242 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -17.13 |
| Consensus MFE | -11.11 |
| Energy contribution | -11.14 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15776144 98 - 20766785 AUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGAAGCUACAUACAUACGUAUCCUAU-------ACUUAGU ((((((((((........(((((....))))).........))))))...((((((((....))-------...)))))).........)))).....-------....... ( -18.33) >DroPse_CAF1 14470 91 - 1 AUAUAUGUAAUAAAUGUUGCAUGUUUACAUGCUUCCAAUACUUGAAUAAUUGGCUUGCUUAUCC-------UAAAAGCCUUA-ACCUACGUAUC-UAU-------A-----U ..(((((((.........(((((....)))))...................(((((........-------...)))))...-...))))))).-...-------.-----. ( -16.20) >DroSec_CAF1 6185 105 - 1 AUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGAAGCUACAUACAUACGUAUCCUAUCCAUCCUACUUAGU ((((((((((........(((((....))))).........))))))...((((((((....))-------...)))))).........))))................... ( -18.33) >DroSim_CAF1 6185 98 - 1 AUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGAAGCUACAUACAUACGUAUCCUAU-------ACUUAGU ((((((((((........(((((....))))).........))))))...((((((((....))-------...)))))).........)))).....-------....... ( -18.33) >DroYak_CAF1 13742 97 - 1 AUACAUUUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGAUUGCUUAUGCUUGCUUUUAAGAGCUACA--------UAUCCUAU-------ACUUAGU ..................(((((....))))).(((((...........)))))..(..((((...((((....))))..))--------))..)...-------....... ( -15.40) >DroPer_CAF1 13305 91 - 1 AUAUAUGUAAUAAAUAUUGCAUGUUUACAUGCUUCCAAUACUUGAAUAAUUGGCUUGCUUAUCC-------UAAAAGCCUUA-ACCUACGUAUC-UAU-------A-----U ..(((((((.........(((((....)))))...................(((((........-------...)))))...-...))))))).-...-------.-----. ( -16.20) >consensus AUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC_______UAAAAGCUACA_ACAUACGUAUCCUAU_______ACUUAGU ....(((((.........(((((....)))))...................(((((..................))))).......)))))..................... (-11.11 = -11.14 + 0.03)
| Location | 15,776,174 – 15,776,274 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -14.01 |
| Energy contribution | -15.40 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15776174 100 + 20766785 UCUA-------GCAUAAGCAAGCCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUACAUGUAUUUACAUACGAUUAAUGCUCUUGAAUGCAGGCA ....-------((....))..(((..(((((.((((((((...(((((....)))))..((.....)))).)))))).)))))......(((........)))))). ( -24.10) >DroSec_CAF1 6222 100 + 1 UCUA-------GCAUAAGCAAGCCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUACAUGUAUUUACAUACGAUUAAUGCUCUUGAAUGCAGGCA ....-------((....))..(((..(((((.((((((((...(((((....)))))..((.....)))).)))))).)))))......(((........)))))). ( -24.10) >DroSim_CAF1 6215 100 + 1 UCUA-------GCAUAAGCAAGCCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUACAUGUAUUUACAUACGAUUAAUGCUCUUGAAUGCAGGCA ....-------((....))..(((..(((((.((((((((...(((((....)))))..((.....)))).)))))).)))))......(((........)))))). ( -24.10) >DroEre_CAF1 7357 93 + 1 CUUAAAAGCAAG----------CCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUACAUAUACUU----ACGAUUAAUGCUCUUGAAUGCAGGCA ...........(----------(((((((((..((((...((.(((((....))))).))...))))...)))))))----........(((........)))))). ( -22.10) >DroYak_CAF1 13764 103 + 1 CUUAAAAGCAAGCAUAAGCAAUCCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUAAAUGUAUUU----ACGAAUUAUGCUCUUGAAUGCAGGCA .......(((..((..((((.(((((.(((....)))))))).(((((....)))))....................----........))))..))..)))..... ( -24.00) >DroPer_CAF1 13328 100 + 1 UUUA-------GGAUAAGCAAGCCAAUUAUUCAAGUAUUGGAAGCAUGUAAACAUGCAAUAUUUAUUACAUAUAUGUAUGUACAAUGUACAAAAUAUGUUCCAGGCA ....-------((((((.........))))))..((.(((((((((((....))))).((((((..((((....))))(((((...))))))))))).)))))))). ( -20.90) >consensus UCUA_______GCAUAAGCAAGCCAAGUAUGCAAGUAUUGGAAGCGUGUAAACACGCAUUAUUUAUUACAUGUAUUUACAUACGAUUAAUGCUCUUGAAUGCAGGCA .....................(((....(((((.(((.((((.(((((....)))))....)))).))).)))))..............(((........)))))). (-14.01 = -15.40 + 1.39)



| Location | 15,776,174 – 15,776,274 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.45 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15776174 100 - 20766785 UGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGA ...((((((....((((.......((((....))))((((((........(((((....))))).........)))))).....))))....))))-------.)). ( -23.73) >DroSec_CAF1 6222 100 - 1 UGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGA ...((((((....((((.......((((....))))((((((........(((((....))))).........)))))).....))))....))))-------.)). ( -23.73) >DroSim_CAF1 6215 100 - 1 UGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC-------UAGA ...((((((....((((.......((((....))))((((((........(((((....))))).........)))))).....))))....))))-------.)). ( -23.73) >DroEre_CAF1 7357 93 - 1 UGCCUGCAUUCAAGAGCAUUAAUCGU----AAGUAUAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGG----------CUUGCUUUUAAG (((((.......)).)))((((..((----((((.(((((((........(((((....))))).........)))))))....)----------)))))..)))). ( -22.53) >DroYak_CAF1 13764 103 - 1 UGCCUGCAUUCAAGAGCAUAAUUCGU----AAAUACAUUUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGAUUGCUUAUGCUUGCUUUUAAG .....(((..((.(((((........----....................(((((....))))).(((((...........))))).))))).))..)))....... ( -21.20) >DroPer_CAF1 13328 100 - 1 UGCCUGGAACAUAUUUUGUACAUUGUACAUACAUAUAUGUAAUAAAUAUUGCAUGUUUACAUGCUUCCAAUACUUGAAUAAUUGGCUUGCUUAUCC-------UAAA .(((((((((((((..(((((...))))).....))))))..........(((((....))))).))))..............)))..........-------.... ( -18.73) >consensus UGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUCCAAUACUUGCAUACUUGGCUUGCUUAUGC_______UAAA .((((((........)))..................((((((........(((((....))))).........))))))....)))..................... (-12.61 = -12.45 + -0.16)

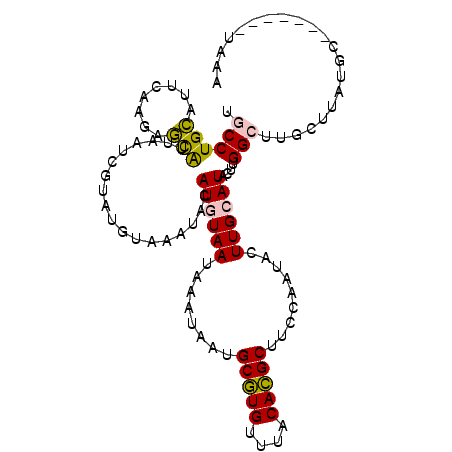
| Location | 15,776,207 – 15,776,304 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -13.13 |
| Energy contribution | -11.17 |
| Covariance contribution | -1.97 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15776207 97 - 20766785 UUUUGCAACAAAGAGCUUUAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUC ..(((((.......(((((....((((((......)))))).))))).......((((....)))).)))))........(((((....)))))... ( -22.40) >DroPse_CAF1 14526 97 - 1 CUUUACAUUAAAUGACUCUUAGCGAGUUUGUGCCUGGAACAUAUUUUGUACAUUGUACAUACAUAUAUGUAAUAAAUGUUGCAUGUUUACAUGCUUC ..((((((...(((((((.....)))).(((((...(.(((.....))).)...)))))..)))..))))))........(((((....)))))... ( -19.70) >DroSim_CAF1 6248 97 - 1 UUUUGCAACAAAGAGCUCUUAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUC ..(((((.......((((((...((((((......)))))))))))).......((((....)))).)))))........(((((....)))))... ( -26.20) >DroEre_CAF1 7387 93 - 1 UUUUGCAACAAAGAGCUCGAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGU----AAGUAUAUGUAAUAAAUAAUGCGUGUUUACACGCUUC ..(((((.......((((.....((((((......))))))..))))......(..----..)....)))))........(((((....)))))... ( -21.20) >DroYak_CAF1 13804 93 - 1 UUUUGCAACAAAGAGCUCGAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUAAUUCGU----AAAUACAUUUAAUAAAUAAUGCGUGUUUACACGCUUC ....((........))..(((((((((.(((((((.......)).)))))))))((----((((((..(((.....)))...))))))))..))))) ( -22.00) >DroPer_CAF1 13361 97 - 1 CUUUACAUUAAAUGACUCUUAGCGAGUUUGUGCCUGGAACAUAUUUUGUACAUUGUACAUACAUAUAUGUAAUAAAUAUUGCAUGUUUACAUGCUUC ..((((((...(((((((.....)))).(((((...(.(((.....))).)...)))))..)))..))))))........(((((....)))))... ( -19.70) >consensus UUUUGCAACAAAGAGCUCUAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUAAAUCGUA__UAAAUACAUGUAAUAAAUAAUGCGUGUUUACACGCUUC ..(((((.((....((((.....))))...))..(((........)))...................)))))........(((((....)))))... (-13.13 = -11.17 + -1.97)


| Location | 15,776,242 – 15,776,341 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15776242 99 - 20766785 A--UAAGUACUCGUUCCGUAUGAGGCACAUAGUCGGGA-GUUUUGCAACAAAGAGCUUUAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAA .--.............(((((((((((((((.((((((-(((((.......))))))))...))).))))))))(((........)))....)))))))... ( -30.60) >DroSec_CAF1 6290 101 - 1 AUAUAAGUACUCGUUCCGUAUGAGGCACAUAGUCGGGA-GUUUUGCAACAAAGAGCUCUAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAA ................(((((((((((((((.((((((-(((((.......))))))))...))).))))))))(((........)))....)))))))... ( -33.30) >DroSim_CAF1 6283 101 - 1 AUAUAAGUACUCGUUCCGUAUGAGGCACAUAGUCGGGA-GUUUUGCAACAAAGAGCUCUUAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAA ................(((((((((((((((.((((((-(((((.......))))))))...))).))))))))(((........)))....)))))))... ( -33.30) >DroEre_CAF1 7422 97 - 1 AUAUAAGUACUCGAUCCGUAUGAGGCACAUAGUCGGGA-GUUUUGCAACAAAGAGCUCGAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGU----AA .....(((.(((.......((((((((((((.(((.((-(((((.......)))))))....))).))))))))).)))....))).)))......----.. ( -29.20) >DroYak_CAF1 13839 97 - 1 AUAUAAGUACUCGAUCCGUAUGAGGCACAUAGUCGGGA-GUUUUGCAACAAAGAGCUCGAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUAAUUCGU----AA .........(((.......((((((((((((.(((.((-(((((.......)))))))....))).))))))))).)))....)))..........----.. ( -28.70) >DroPer_CAF1 13396 102 - 1 AUACAUUUAUAUGCAUCGUAUCAGGCACAUAUUCGGCACGCUUUACAUUAAAUGACUCUUAGCGAGUUUGUGCCUGGAACAUAUUUUGUACAUUGUACAUAC .................((.(((((((((.....(((.((((..................)))).)))))))))))).))......(((((...)))))... ( -22.87) >consensus AUAUAAGUACUCGUUCCGUAUGAGGCACAUAGUCGGGA_GUUUUGCAACAAAGAGCUCUAAGCGAGUGUGUGCCUGCAUUCAAGAGCAUUAAUCGUAUGUAA .....(((.(((.......((((((((((((.((((((.(((((.......))))))))...))).))))))))).)))....))).)))............ (-20.83 = -21.50 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:40 2006