| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,775,888 – 15,775,990 |
| Length | 102 |
| Max. P | 0.726077 |

| Location | 15,775,888 – 15,775,990 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
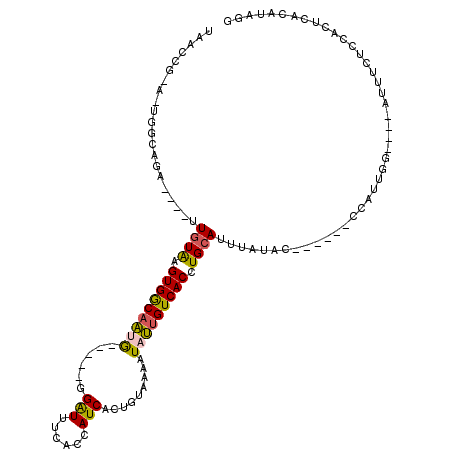
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -11.02 |
| Energy contribution | -10.74 |
| Covariance contribution | -0.28 |
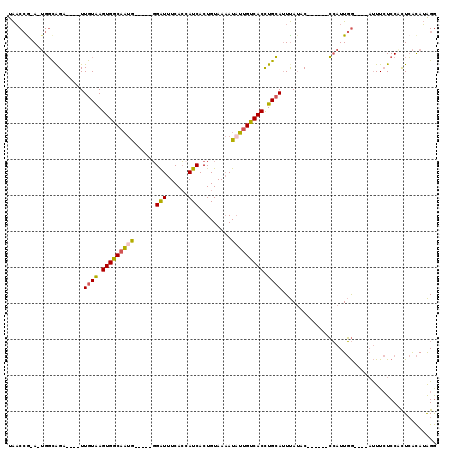
| Combinations/Pair | 1.31 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
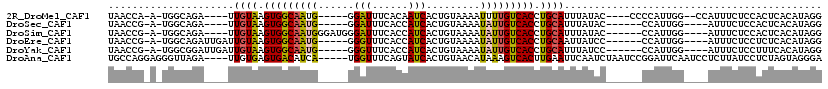
>2R_DroMel_CAF1 15775888 102 + 20766785 UAACCA-A-UGGCAGA----UUGUAAGUGGCAAUG-----GGAUUUCACAAUCACUGUAAAAUUUUGUCACCUGCAUUUAUAC----CCCCAUUGG--CCAUUUCUCCACUCACAUAGG ...(((-(-(((....----.((((.(((((((..-----.(((((.(((.....))).)))))))))))).)))).......----..)))))))--..................... ( -22.14) >DroSec_CAF1 5937 98 + 1 UAACCG-A-UGGCAGA----UUGUAAGUGGCAAUG-----GGAUUUCACCAUCACUGUAAAAUAUUGUCACCUGCAUUUAUAC------CCAUUGG----AUUUCUCCACUCACAUAGG ...(((-(-(((..((----.((((.(((((((((-----......((.......)).....))))))))).)))).))....------)))))))----................... ( -22.80) >DroSim_CAF1 5932 103 + 1 UAACCG-A-UGGCAGA----UUGUAAGUGGCAAUGGGAUGGGAUUUCACCAUCACUGUAAAAUAUUGUCACCUGCAUUUAUAC------CCAUUGG----AUUUCUCCACUCACAUAGG ...(((-(-(((..((----.((((.(((((((((.(((((.......))))).........))))))))).)))).))....------)))))))----................... ( -29.80) >DroEre_CAF1 7086 102 + 1 UAACCG-A-UGGCAGAUUGAUUGUAAGUGGCAAUG-----GGGUUUCACCAUCACUGUAAAAUAUUGUCACCUGCAAUUAUCC------CCAUUGG----AUUUCUCCUCUCACAUAGG ...(((-(-(((..(((.(((((((.(((((((((-----.((.....))............))))))))).)))))))))).------)))))))----................... ( -29.12) >DroYak_CAF1 13493 102 + 1 UAACCG-A-UGGCGGAUUGAUUGUAAGUGGCAAUG-----GGGUUUCACCAUCACUGUAAAAUAUUGUCACCUGCAUUUAUCC------CCAUUGG----AUUUCUCCUUUCACAUAGG ...(((-(-(((.((((.((.((((.(((((((((-----.((.....))............))))))))).)))).))))))------)))))))----......(((.......))) ( -31.32) >DroAna_CAF1 5633 110 + 1 UGCCAGGAGGGUUAGA----UUGUGAGUGACAUCA-----UGGUUUCAGUAUCACUGUAACAUAAAGUCACUUGAAUUCAAUCUAAUCCGGAUUCAAUCCUCUUAUCCUCUAGUAGGGA (((.((((((((((((----(((.(((((((....-----..(((.(((.....))).))).....))))))).....)))))))))))((((...)))).....))))...))).... ( -34.00) >consensus UAACCG_A_UGGCAGA____UUGUAAGUGGCAAUG_____GGAUUUCACCAUCACUGUAAAAUAUUGUCACCUGCAUUUAUAC______CCAUUGG____AUUUCUCCACUCACAUAGG .....................((((.(((((((((......(((......))).........))))))))).))))........................................... (-11.02 = -10.74 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:36 2006