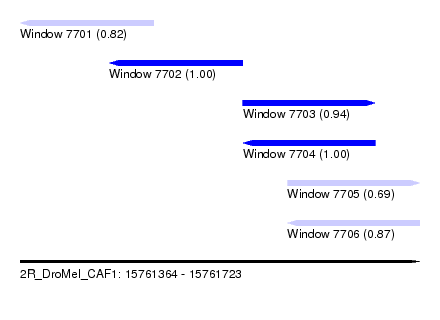
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,761,364 – 15,761,723 |
| Length | 359 |
| Max. P | 0.999678 |

| Location | 15,761,364 – 15,761,484 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -48.42 |
| Consensus MFE | -44.38 |
| Energy contribution | -43.58 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761364 120 - 20766785 ACGUGACUGAGGCUGCUGCCUCGGUGACCAUUACCGCUCAAUCCGAAAGCUUUGUGGAGGUAUCGCCCACGGUGCAGAGCUCCUCGGUGGAGGUGCUGGAACUUUCGGAACGUAAGAGGG ..((.((((((((....)))))))).)).....((......(((((((((.((((((.(......))))))).))..(((.((((....)))).))).....))))))).(....).)). ( -47.70) >DroSec_CAF1 9839 120 - 1 AUGUGACUGAGGCGGCUACUUCGGUAACAAUUACUGCUCAAUCCGAAAGCUUUGUGGAGGUAUCGCCGACGGUGCAGAGCUCCUCGGUGGAGGUGCUAGAACUUUCGGAACGUAAGAGGG .(((.(((((((......))))))).))).......(((..((((((((.(((......((((((....))))))..(((.((((....)))).)))))).)))))))).(....).))) ( -38.80) >DroSim_CAF1 10717 120 - 1 ACGUGACCGAGGCUGCUGCCUCGGUGACCAUUACCGCUCAAUCCGAGAGCUUUGUGGAGGUAUCGCCGACGGUGCAGAGCUCCUCGGUGGAGGUGCUGGAACUUUCGGAACGUAAGAGGG ..((.((((((((....)))))))).)).....((.(((...((((((((((((((..((.....))..)...)))))))).)))))(((((((......)))))))........))))) ( -51.40) >DroEre_CAF1 10414 120 - 1 ACGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCAAUCUGAGAGCUUUGUGGAGGUAUCGCCCACGGUGCAGAGCUCCUCGGUGGAGGUGCUGGAGCUUUCGGAACGUAAGAGGG ..((.((((((((....)))))))).)).....((......((((((((((((......((((((....))))))..(((.((((....)))).))))))))))))))).(....).)). ( -50.90) >DroYak_CAF1 10553 120 - 1 AUGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCAGUCUGAGAGCUUUGUGGAGGUCUCGCCCACGGUGCAGAGCUCCUCGGCGGAGGUGCUGGAGCUUUCGGAGCGUAAGAGGG ..((.((((((((....)))))))).)).....((.(((.((((((((((((...((((.(((((((...)))).))).)))).(((((....)))))))))))))))).)....))))) ( -53.30) >consensus ACGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCAAUCCGAGAGCUUUGUGGAGGUAUCGCCCACGGUGCAGAGCUCCUCGGUGGAGGUGCUGGAACUUUCGGAACGUAAGAGGG ..((.((((((((....)))))))).)).....((......((((((((.(((......((((((....))))))..(((.((((....)))).))).))))))))))).(....).)). (-44.38 = -43.58 + -0.80)

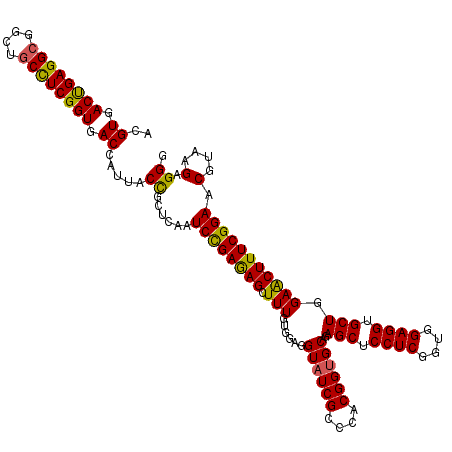
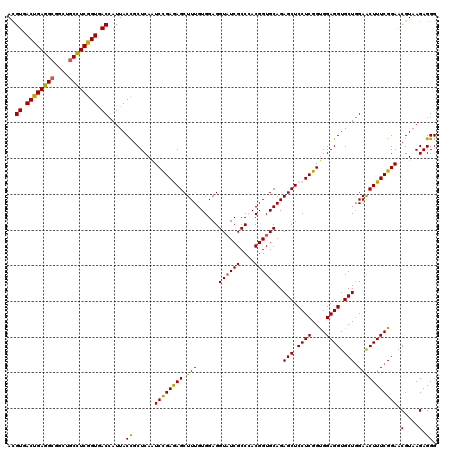
| Location | 15,761,444 – 15,761,564 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -49.46 |
| Consensus MFE | -46.14 |
| Energy contribution | -45.74 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761444 120 - 20766785 ACUGACCUUCGUGGGUGCGGAGGGCAGCGAGCGGAACUUAAACACUGGCCUGAUAGUACUGGUUCACCAUCCCAUGGACUACGUGACUGAGGCUGCUGCCUCGGUGACCAUUACCGCUCA .(((.(((((((....))))))).))).((((((.........((((......))))..((((.((((....((((.....))))...(((((....)))))))))))))...)))))). ( -50.60) >DroSec_CAF1 9919 120 - 1 ACUGACCUUCGUGGGUGCGGAGGGCAGCGAGCGGAACUUAAACACCGGCCUGAUAGUACUGGUCCACCAUCCCAUGGACUAUGUGACUGAGGCGGCUACUUCGGUAACAAUUACUGCUCA .(((.(((((((....))))))).))).((((((..........(((.(((..(((((((((((((........))))))).)).)))))))))).(((....))).......)))))). ( -45.10) >DroSim_CAF1 10797 120 - 1 ACUGACCUUCGUGGGUGCGGAGGGCAGCGAGCGUAACUUAAACACCGGCCUGAUAGUCCUGGUCCACCAUCCCAUGGACUACGUGACCGAGGCUGCUGCCUCGGUGACCAUUACCGCUCA .(((.(((((((....))))))).))).((((((((.................(((((((((.........))).)))))).((.((((((((....)))))))).))..))).))))). ( -50.60) >DroEre_CAF1 10494 120 - 1 UCUGACCUUCGUGGGUGCGGAGGGCAGCGAGCGGAACCUAAACACCGGCCUGAUUGUCCUGGUCCACCACCCCAUGGACUACGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCA .(((.(((((((....))))))).))).((((((............((.(.....).))(((((((........))))))).((.((((((((....)))))))).)).....)))))). ( -49.30) >DroYak_CAF1 10633 120 - 1 ACUGACCUUUGUGGGUGCGGAGGGCAGUGAGCGGAAUUUAAACACUGGCCUUAUAGUGCUGGUCCACCAUCCCAUGGACUAUGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCA ((((.(((((((....))))))).))))((((((........(((((......))))).(((((((........))))))).((.((((((((....)))))))).)).....)))))). ( -51.70) >consensus ACUGACCUUCGUGGGUGCGGAGGGCAGCGAGCGGAACUUAAACACCGGCCUGAUAGUACUGGUCCACCAUCCCAUGGACUACGUGACUGAGGCGGCUGCCUCGGUGACCAUUACCGCUCA .(((.(((((((....))))))).))).((((((.........................(((((((........))))))).((.((((((((....)))))))).)).....)))))). (-46.14 = -45.74 + -0.40)


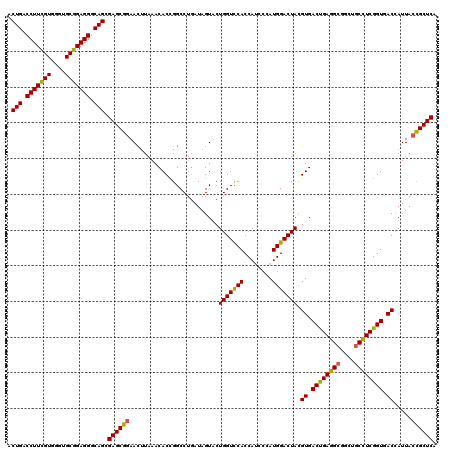
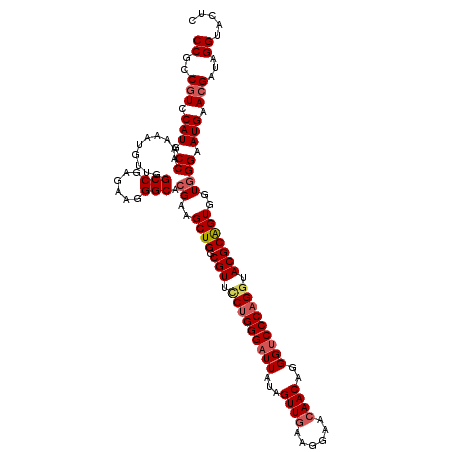
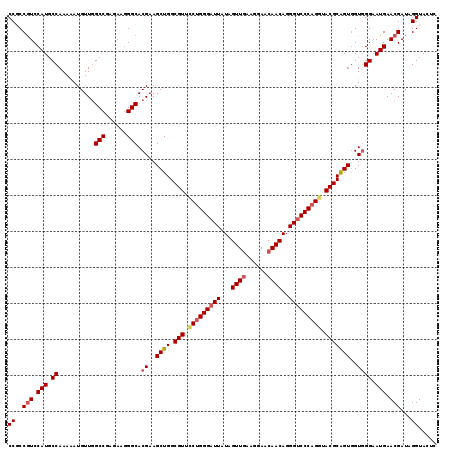
| Location | 15,761,564 – 15,761,683 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -36.22 |
| Energy contribution | -37.14 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761564 119 + 20766785 CCGCCAUCCAUCCCAAAAAUGUUGGCCGAGAAGGGCACGAAGCUGGCGUUCCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGAUACGCAGUGGUGGGAAUGAACGAUAGGUACUC ..(((((((((((((......((((((......))).))).((((.(((..((((((((...((((......))))..))))))))..)))))))..)))).)))...))).))).... ( -41.40) >DroSec_CAF1 10039 119 + 1 CCUCCGUCCAUGCCAAAAAUGUUAGCCGAGAAGGGCACGAAGCUGGCGUUUCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUGGUGGGUAUGAACGAUAGGUACUC (((.(((.((((((..........(((......))).((..((((.(((.(((((((((...((((......))))..))))))))).)))))))..)))))))).)))..)))..... ( -45.90) >DroSim_CAF1 10917 119 + 1 CCUCCGUCCAUGCCAAAGAUGUUGGCCGAGAAAGGCACGAAGCUGGCGUUCCUGGGAUUGUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUUGUGGGAAUGAACGAUAGGUACUC (((.(((.(((.(((......((((((......))).)))(((((.(((.(((((((((.(.((((......))))).))))))))).)))))))).)))..))).)))..)))..... ( -44.50) >DroEre_CAF1 10614 119 + 1 CCGCCGUCCAUGCCAAAUAUGUUGGCCGAGAAGGGCACGAAGCUGGCGUUCCUGGGAUUAUAGUUAAAGGAGCAACAGGGACCCAGGUACGCGGUGGUUGGAAUGAACGAUAGGUACUC (((((((((..(((((.....)))))......)))).........((((.((((((.((...(((........)))...)))))))).)))))))))...................... ( -36.50) >DroYak_CAF1 10753 119 + 1 CCGCCGUCCAUGCCAAAGAUGUUGGCCGAGAAGGGCACGAAGCUGGCGUUCCGGGGAUUAUAGUUGAAGGAGCAACAGGGUCCCAGGUACGCAGUGGUGGGAAUGAACGAUAGGUACUC (((((((((..(((((.....)))))......)))).....((((.(((.((.((((((...((((......))))..)))))).)).))))))))))))................... ( -41.60) >consensus CCGCCGUCCAUGCCAAAAAUGUUGGCCGAGAAGGGCACGAAGCUGGCGUUCCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUGGUGGGAAUGAACGAUAGGUACUC ((..(((.(((.((..........(((......))).((..((((.(((.(((((((((...((((......))))..))))))))).)))))))..)))).))).)))...))..... (-36.22 = -37.14 + 0.92)

| Location | 15,761,564 – 15,761,683 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -34.04 |
| Energy contribution | -34.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761564 119 - 20766785 GAGUACCUAUCGUUCAUUCCCACCACUGCGUAUCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGGAACGCCAGCUUCGUGCCCUUCUCGGCCAACAUUUUUGGGAUGGAUGGCGG ......((..(((((((.((((...((((((.(((((((....((((......)))).....))))))).))).)))....(.(((......))))........)))))))))))..)) ( -39.10) >DroSec_CAF1 10039 119 - 1 GAGUACCUAUCGUUCAUACCCACCACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGAAACGCCAGCUUCGUGCCCUUCUCGGCUAACAUUUUUGGCAUGGACGGAGG .....(((.((((((((.((.....((((((..((((((....((((......)))).....))))))..))).)))......(((......)))..........)).))))))))))) ( -36.40) >DroSim_CAF1 10917 119 - 1 GAGUACCUAUCGUUCAUUCCCACAACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUACAAUCCCAGGAACGCCAGCUUCGUGCCUUUCUCGGCCAACAUCUUUGGCAUGGACGGAGG .....(((.((((((((...(((((((((((.(((((((....((((......)))).....))))))).))).))).)).))).........(((((.....)))))))))))))))) ( -41.60) >DroEre_CAF1 10614 119 - 1 GAGUACCUAUCGUUCAUUCCAACCACCGCGUACCUGGGUCCCUGUUGCUCCUUUAACUAUAAUCCCAGGAACGCCAGCUUCGUGCCCUUCUCGGCCAACAUAUUUGGCAUGGACGGCGG ......((..(((((((.((((.....((((.((((((.....((((......))))......)))))).)))).......(.(((......)))).......)))).)))))))..)) ( -37.40) >DroYak_CAF1 10753 119 - 1 GAGUACCUAUCGUUCAUUCCCACCACUGCGUACCUGGGACCCUGUUGCUCCUUCAACUAUAAUCCCCGGAACGCCAGCUUCGUGCCCUUCUCGGCCAACAUCUUUGGCAUGGACGGCGG ......((..(((((((...(((..((((((.((.((((....((((......)))).....)))).)).))).)))....))).........(((((.....))))))))))))..)) ( -33.50) >consensus GAGUACCUAUCGUUCAUUCCCACCACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGGAACGCCAGCUUCGUGCCCUUCUCGGCCAACAUCUUUGGCAUGGACGGCGG ......((..(((((((.((.......((((.(((((((....((((......)))).....))))))).)))).......(.(((......)))).........)).)))))))..)) (-34.04 = -34.04 + -0.00)



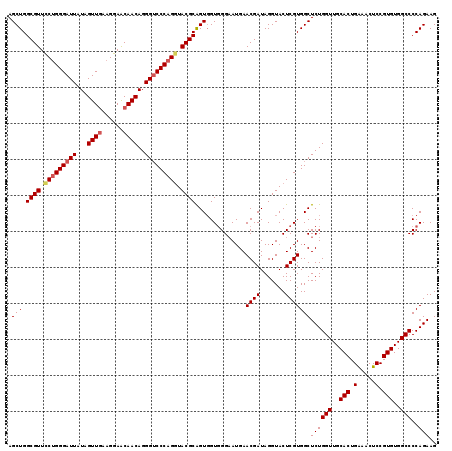
| Location | 15,761,604 – 15,761,723 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.63 |
| Mean single sequence MFE | -42.96 |
| Consensus MFE | -37.58 |
| Energy contribution | -38.26 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761604 119 + 20766785 AGCUGGCGUUCCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGAUACGCAGUGGUGGGAAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGAAACUCCGUGUGGCCCCAGAAG .((((.(((..((((((((...((((......))))..))))))))..)))))))((.(((.....((((.......))))..))).(((..(((.(.....).)))..)))))..... ( -40.70) >DroSec_CAF1 10079 119 + 1 AGCUGGCGUUUCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUGGUGGGUAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGAAACUCCGUGUGGCCCCAGAAG ..(((((((.(((((((((...((((......))))..))))))))).))))(((..(((((((.........)))))))..)))..(((..(((.(.....).)))..))).)))... ( -44.20) >DroSim_CAF1 10957 119 + 1 AGCUGGCGUUCCUGGGAUUGUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUUGUGGGAAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGAAACUCCGUGUGGCCCCAGAAG ..(((((((.(((((((((.(.((((......))))).))))))))).))))(((..((((.((.........)).))))..)))..(((..(((.(.....).)))..))).)))... ( -45.80) >DroEre_CAF1 10654 119 + 1 AGCUGGCGUUCCUGGGAUUAUAGUUAAAGGAGCAACAGGGACCCAGGUACGCGGUGGUUGGAAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGGAACUCCGUGUGGCCCCAGAAG ..((((.((((((..(((....)))..))))))..(((((.(((..((((.(.((.(((......))).)).)))))..).)))))))((..(((.((....)))))..)).))))... ( -42.30) >DroYak_CAF1 10793 119 + 1 AGCUGGCGUUCCGGGGAUUAUAGUUGAAGGAGCAACAGGGUCCCAGGUACGCAGUGGUGGGAAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGGAACUCCGUGUGGCCCCAGAAG .((((.(((.((.((((((...((((......))))..)))))).)).)))))))((.(((.....((((.......))))..))).(((..(((.((....)))))..)))))..... ( -41.80) >consensus AGCUGGCGUUCCUGGGAUUAUAGUUGAAGGAACAACAGGGUCCCAGGUACGCAGUGGUGGGAAUGAACGAUAGGUACUCGUGGCUCUGGUUGCACUGAAACUCCGUGUGGCCCCAGAAG .(((.((((.(((((((((...((((......))))..))))))))).))))..............((((.......)))))))((((((..(((.(.....).)))..)))..))).. (-37.58 = -38.26 + 0.68)


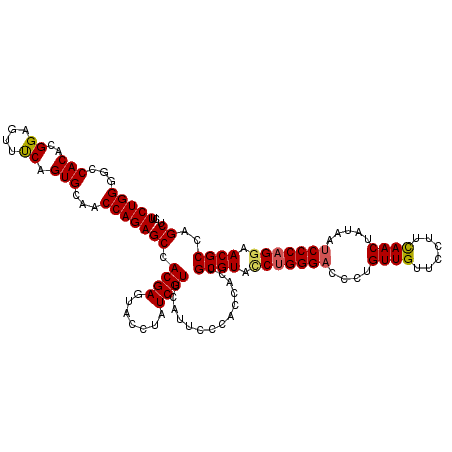
| Location | 15,761,604 – 15,761,723 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.63 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15761604 119 - 20766785 CUUCUGGGGCCACACGGAGUUUCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUUCCCACCACUGCGUAUCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGGAACGCCAGCU ..(((((...(((...........)))...)))))((.((((.......))))..............((((.(((((((....((((......)))).....))))))).))))..)). ( -32.20) >DroSec_CAF1 10079 119 - 1 CUUCUGGGGCCACACGGAGUUUCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUACCCACCACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGAAACGCCAGCU ..(((((...(((...........)))...)))))((.((((.......))))..............((((..((((((....((((......)))).....))))))..))))..)). ( -30.60) >DroSim_CAF1 10957 119 - 1 CUUCUGGGGCCACACGGAGUUUCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUUCCCACAACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUACAAUCCCAGGAACGCCAGCU ..(((((...(((...........)))...)))))((.((((.......))))..............((((.(((((((....((((......)))).....))))))).))))..)). ( -34.60) >DroEre_CAF1 10654 119 - 1 CUUCUGGGGCCACACGGAGUUCCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUUCCAACCACCGCGUACCUGGGUCCCUGUUGCUCCUUUAACUAUAAUCCCAGGAACGCCAGCU ..(((((...(((..((....)).)))...)))))((.((((.......))))..............((((.((((((.....((((......))))......)))))).))))..)). ( -34.00) >DroYak_CAF1 10793 119 - 1 CUUCUGGGGCCACACGGAGUUCCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUUCCCACCACUGCGUACCUGGGACCCUGUUGCUCCUUCAACUAUAAUCCCCGGAACGCCAGCU ..(((((...(((..((....)).)))...)))))((.((((.......))))..............((((.((.((((....((((......)))).....)))).)).))))..)). ( -31.70) >consensus CUUCUGGGGCCACACGGAGUUUCAGUGCAACCAGAGCCACGAGUACCUAUCGUUCAUUCCCACCACUGCGUACCUGGGACCCUGUUGUUCCUUCAACUAUAAUCCCAGGAACGCCAGCU ..(((((...(((..((....)).)))...)))))((.((((.......))))..............((((.(((((((....((((......)))).....))))))).))))..)). (-31.82 = -31.90 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:27 2006