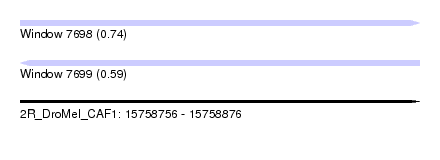
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,758,756 – 15,758,876 |
| Length | 120 |
| Max. P | 0.739348 |

| Location | 15,758,756 – 15,758,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -27.87 |
| Energy contribution | -28.85 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
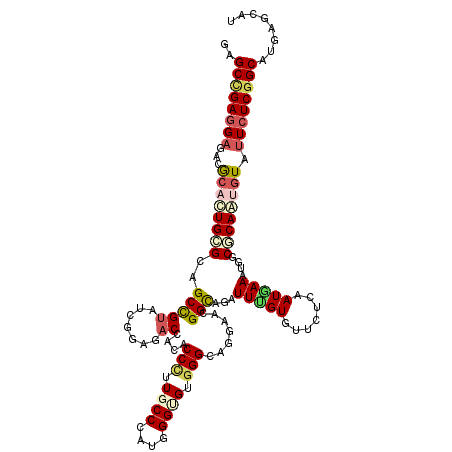
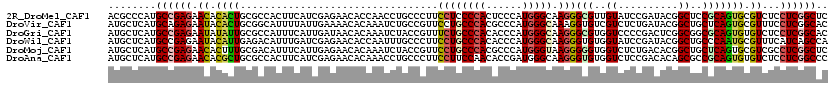
>2R_DroMel_CAF1 15758756 120 + 20766785 GAGCCGAGGAGACGCACUGCGGAGCCGUAUCGGAUACAACGCCCUUGCCCAUGGGAGUGGGGAGGAAGGGCAGGUUGGUGUUCUCGAUGAAGUGGCGCAGUGUGUUCUCGGCAUGGGCGU ..((((((..(((((((((((..((..(((((((..((((((((((.(((((....)))))....))))))..))))....)).)))))..))..)))))))))))))))))........ ( -58.60) >DroVir_CAF1 9386 120 + 1 GUGCCGAGGAAACGCACUGAGCAGCCGUAUCAGAGACGACACCUUUGCCCAUGGGCGUGGGCAGGAACGGCAGAUUUGUGUUUUCAAUAAAAUGCCGCAGUGUAUUCUCUGCAUGAGCAU ((((...(....)))))......(((((..((((((..((((.(((((((((....)))))))))..(((((..(((((.......))))).)))))..))))..)))))).))).)).. ( -43.70) >DroGri_CAF1 6327 120 + 1 GUGCCGAGGAGACACACUGCGCCGCCGAGUCGGGGACCACGCCCUUGCCCAUGGGUGUGGGCAGAAACGGUAGAUUUGUGUUAUCAAUGAAAUGGCGCAAUAUAUUCUCGGCAUGAGCAU ((((((((((.......((((((((((..((...(.((((((((........)))))))).).))..)))).(((.......)))........)))))).....))))))))))...... ( -51.90) >DroWil_CAF1 12413 120 + 1 UGGCUGAUGAAACGCAUUGGGCAGCCGUAUCGGAUACCACACCCUUGCCCAUGGGUGUGGGCAGGAAGGGCAAAUUGGUGUUCUCGAUCAAAUGUCUCAAUGUAUUCUCGGCAUGAGCAU ..(((((.(((..((((((((((...(.(((((((((((..(((((((((((....)))))))...)))).....))))).)).)))))...)))).)))))).))))))))........ ( -45.00) >DroMoj_CAF1 9417 120 + 1 GAGCCGAGGCGACGCACUGAGCAGCCGUGUCAGAGACCACCCCCUUACCCAUGGGCGUGGGCAGGAACGGUAGAUUUGUGUUCUCAAUGAAAUGUCGCAAAGUGUUCUCGGCAUGAGCAU ..(((((((....(((((..((.(((((.((.....((((.(((........))).))))....))))))).(((.....(((.....)))..)))))..))))))))))))........ ( -43.00) >DroAna_CAF1 7788 120 + 1 GGGCCGAGGAGACACACUGCGGCGCUGUGUCGGAGACCACACCCUUGCCCAUCGGUGUUGGAAGGAAGGGCAGGUUUGUGUUCUCGAUGAAGUGGCGCAGCGUGUUCUCGGCAUGAGCAU ..((((((..(((((.(((((.((((.(((((.(((.((((..(((((((.((...........)).)))))))..)))).)))))))).)))).))))).)))))))))))........ ( -61.40) >consensus GAGCCGAGGAGACGCACUGCGCAGCCGUAUCGGAGACCACACCCUUGCCCAUGGGUGUGGGCAGGAACGGCAGAUUUGUGUUCUCAAUGAAAUGGCGCAAUGUAUUCUCGGCAUGAGCAU ..((((((((...((((((((..(((((.......))....(((.((((....)))).))).......)))...(((((.......)))))....)))))))).))))))))........ (-27.87 = -28.85 + 0.98)

| Location | 15,758,756 – 15,758,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15758756 120 - 20766785 ACGCCCAUGCCGAGAACACACUGCGCCACUUCAUCGAGAACACCAACCUGCCCUUCCUCCCCACUCCCAUGGGCAAGGGCGUUGUAUCCGAUACGGCUCCGCAGUGCGUCUCCUCGGCUC ........((((((.((.(((((((...((..((((.((....((((..((((((....((((......)))).))))))))))..))))))..))...))))))).))...)))))).. ( -44.50) >DroVir_CAF1 9386 120 - 1 AUGCUCAUGCAGAGAAUACACUGCGGCAUUUUAUUGAAAACACAAAUCUGCCGUUCCUGCCCACGCCCAUGGGCAAAGGUGUCGUCUCUGAUACGGCUGCUCAGUGCGUUUCCUCGGCAC ..(((....((((((..(((((((((((..((..((....))..))..))))))...((((((......))))))..)))))..))))))....)))((((.((........)).)))). ( -38.50) >DroGri_CAF1 6327 120 - 1 AUGCUCAUGCCGAGAAUAUAUUGCGCCAUUUCAUUGAUAACACAAAUCUACCGUUUCUGCCCACACCCAUGGGCAAGGGCGUGGUCCCCGACUCGGCGGCGCAGUGUGUCUCCUCGGCAC .......(((((((.((((((((((((......(((......))).....(((..((((((((......)))))).(((.(....))))))..))).))))))))))))...))))))). ( -49.00) >DroWil_CAF1 12413 120 - 1 AUGCUCAUGCCGAGAAUACAUUGAGACAUUUGAUCGAGAACACCAAUUUGCCCUUCCUGCCCACACCCAUGGGCAAGGGUGUGGUAUCCGAUACGGCUGCCCAAUGCGUUUCAUCAGCCA ..(((.(((....(((..(((((.(.((.(((((((.((..((((....(((((...((((((......))))))))))).)))).)))))).))).))).)))))..)))))).))).. ( -36.80) >DroMoj_CAF1 9417 120 - 1 AUGCUCAUGCCGAGAACACUUUGCGACAUUUCAUUGAGAACACAAAUCUACCGUUCCUGCCCACGCCCAUGGGUAAGGGGGUGGUCUCUGACACGGCUGCUCAGUGCGUCGCCUCGGCUC ........((((((........(((((....(((((((........((.((((..((((((((......)))))..)))..))))....))........))))))).))))))))))).. ( -41.89) >DroAna_CAF1 7788 120 - 1 AUGCUCAUGCCGAGAACACGCUGCGCCACUUCAUCGAGAACACAAACCUGCCCUUCCUUCCAACACCGAUGGGCAAGGGUGUGGUCUCCGACACAGCGCCGCAGUGUGUCUCCUCGGCCC ........((((((.((((((((((.(.((...((((((.((((..(.(((((.((...........)).))))).)..)))).))).)))...)).).))))))))))...)))))).. ( -51.00) >consensus AUGCUCAUGCCGAGAACACACUGCGCCAUUUCAUCGAGAACACAAAUCUGCCCUUCCUGCCCACACCCAUGGGCAAGGGUGUGGUCUCCGACACGGCUGCGCAGUGCGUCUCCUCGGCAC ........((((((.((.((((.................................((((((((......))))).)))(((..((..........))..))))))).))...)))))).. (-21.33 = -21.67 + 0.34)
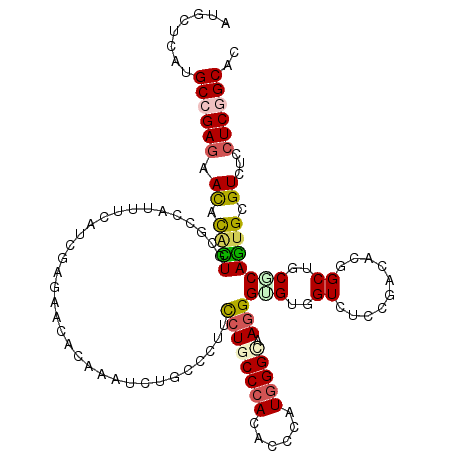
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:20 2006