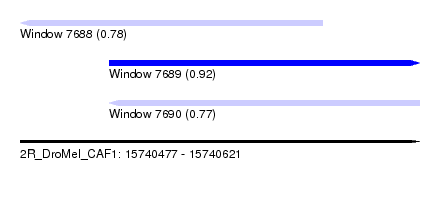
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,740,477 – 15,740,621 |
| Length | 144 |
| Max. P | 0.919919 |

| Location | 15,740,477 – 15,740,586 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15740477 109 - 20766785 AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCGGAUAAACAAAUAUUCAAUUCUUUGGCUUCUU ...((((.((((((((((.(....).)))))))))).).)))..((((.((((.((((((.......))))))....(((((......)))))..))))))))...... ( -28.70) >DroSec_CAF1 616278 109 - 1 AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCGGAUAAGCAAAUAUUCAAUUCUUUGGCUCCCU ........((((((((((.(....).))))))))))(((.((((.....((((.((((((.......))))))....(((((......)))))..))))..))))))). ( -32.60) >DroSim_CAF1 523822 107 - 1 AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCGGAUAAGCAAAUAUUCAAUUCUU--GCUCCUU .....(..(..(((((((((((....))).(((((((((....))).))))))....))))))))..)..)......(((...((((...........))--))))).. ( -28.10) >DroEre_CAF1 520855 108 - 1 AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGUGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCCGAUAAACAAAUAUUCCAUUCUUUGGCC-CUU ........((((((((((.(....).))))))))))(((.((((......(((.((((((.......))))))..)))((((......)))).........))))-))) ( -29.20) >DroYak_CAF1 539669 107 - 1 AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGUGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCGGAUAAAUAAAUGUUCAAUUCGUUGGCC--UU ......(.((((((((((.(....).)))))))))).)..((((......((..((((((.......))))))..))(((((......)))))........))))--.. ( -29.40) >consensus AUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUCGGAUAAACAAAUAUUCAAUUCUUUGGCU_CUU ........((((((((((.(....).))))))))))(((.((((((....))..((((((.......))))))....(((((......)))))........)))).))) (-26.66 = -26.70 + 0.04)

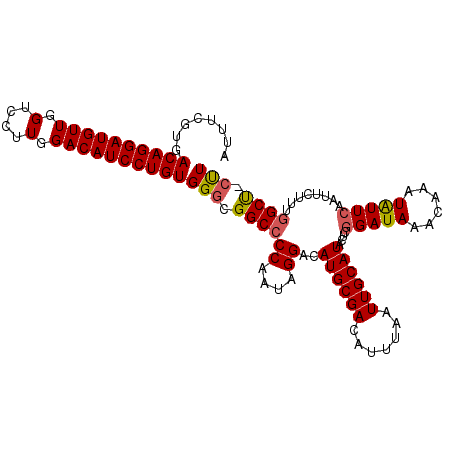
| Location | 15,740,509 – 15,740,621 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -32.62 |
| Energy contribution | -33.22 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15740509 112 + 20766785 GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCGCCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCAGUCAUGACCUGCUGCACCAUUGGGAACCACA ...(((((.((.....)).)))))(((((.(((.((.((..(((((((((..........)))))))))........((((....))))..)).)).))).)))))...... ( -33.20) >DroSec_CAF1 616310 112 + 1 GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCGCCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCAGUCGUGACCUGCUGCACCAUUGGGAACCACA ...(((((.((.....)).)))))(((((.(((.((.((....(((((((..........)))))))(((((((........)))))))..)).)).))).)))))...... ( -35.70) >DroSim_CAF1 523852 112 + 1 GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCGCCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCAGUCGUGACCUGCUGCACCAUUGGGAACCACG ...(((((.((.....)).)))))(((((.(((.((.((....(((((((..........)))))))(((((((........)))))))..)).)).))).)))))...... ( -35.70) >DroEre_CAF1 520886 112 + 1 GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCACCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCAGUCGUGACCUGCUGCACCAUUGGGAACCACA ...(((((.((.....)).)))))(((((.(((.((.......(((((((..........)))))))(((((((........))))))).....)).))).)))))...... ( -32.70) >DroYak_CAF1 539699 112 + 1 GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCACCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCACUCGUGACCAGCUGCACCAUUGGGAACCACA ...(((((.((.....)).)))))(((((.(((.((.......(((((((..........)))))))(((((((........))))))).....)).))).)))))...... ( -32.90) >consensus GAGUAUGCAAUUAAAUGUCGCAUGUCCUAUUGGGGCCGCCCACAGGAUGUCCAAGGACCAACAUCCUGUCACGAAAUGUCAGUCGUGACCUGCUGCACCAUUGGGAACCACA ...(((((.((.....)).)))))(((((.(((.((.((....(((((((..........)))))))(((((((........)))))))..)).)).))).)))))...... (-32.62 = -33.22 + 0.60)

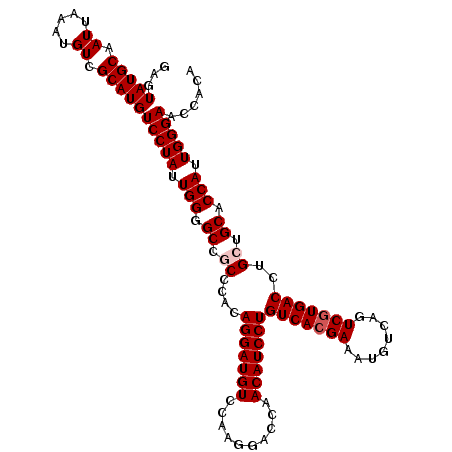
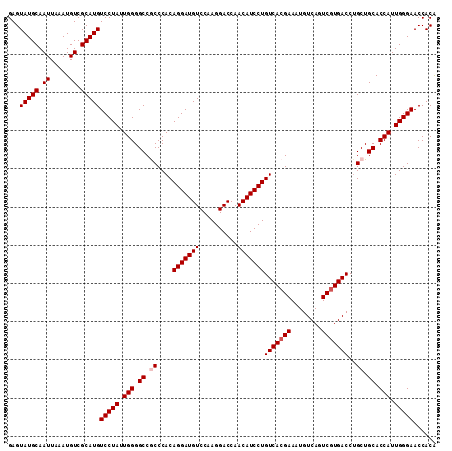
| Location | 15,740,509 – 15,740,621 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -37.34 |
| Energy contribution | -37.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
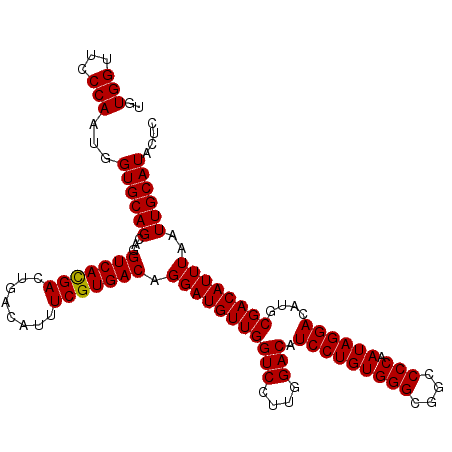
>2R_DroMel_CAF1 15740509 112 - 20766785 UGUGGUUCCCAAUGGUGCAGCAGGUCAUGACUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC ..(((...)))...((((((...(((((((........))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))))).... ( -35.70) >DroSec_CAF1 616310 112 - 1 UGUGGUUCCCAAUGGUGCAGCAGGUCACGACUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC ..(((...)))...((((((...(((((((........))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))))).... ( -37.50) >DroSim_CAF1 523852 112 - 1 CGUGGUUCCCAAUGGUGCAGCAGGUCACGACUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC .(((((.((....)).)).(((((((((((........))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))).))).. ( -37.70) >DroEre_CAF1 520886 112 - 1 UGUGGUUCCCAAUGGUGCAGCAGGUCACGACUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGUGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC ..(((...)))...((((((...(((((((........))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))))).... ( -37.80) >DroYak_CAF1 539699 112 - 1 UGUGGUUCCCAAUGGUGCAGCUGGUCACGAGUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGUGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC ..(((...)))...((((((...((((((((......)))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))))).... ( -39.50) >consensus UGUGGUUCCCAAUGGUGCAGCAGGUCACGACUGACAUUUCGUGACAGGAUGUUGGUCCUUGGACAUCCUGUGGGCGGCCCCAAUAGGACAUGCGACAUUUAAUUGCAUACUC ..(((...)))...((((((...(((((((........))))))).(((((((((((....))).(((((((((....))).))))))....))))))))..)))))).... (-37.34 = -37.18 + -0.16)

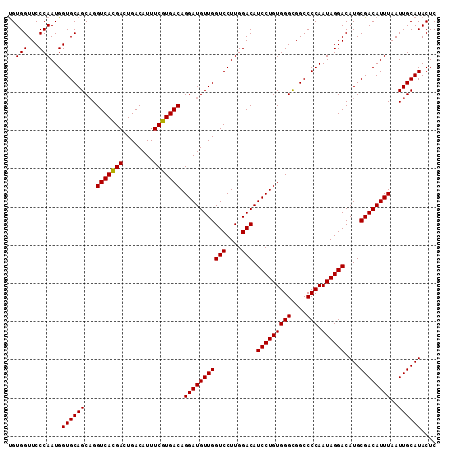
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:11 2006