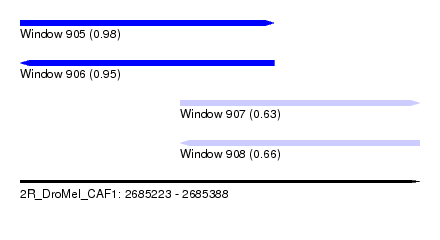
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,685,223 – 2,685,388 |
| Length | 165 |
| Max. P | 0.983431 |

| Location | 2,685,223 – 2,685,328 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -15.68 |
| Energy contribution | -17.65 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
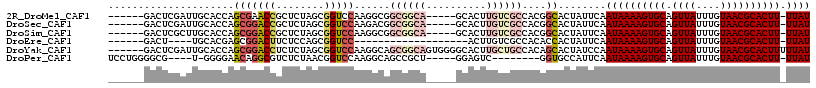
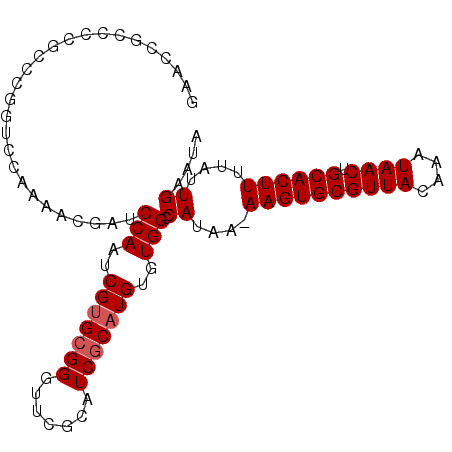
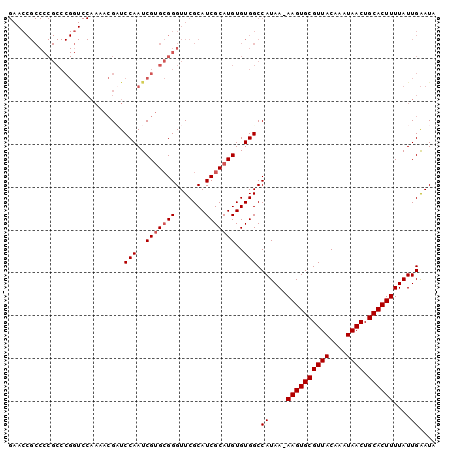
>2R_DroMel_CAF1 2685223 105 + 20766785 ------GACUCGAUUGCACCAGCGAACCGCUCUAGCGGUCCAAGGCGGCGGCA-----GCACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAU ------.....((.(((......(.(((((....))))).)..((((((((..-----....))))))))...)))....)).((((((((((.((((....))))))))))-)))) ( -35.70) >DroSec_CAF1 117825 105 + 1 ------GACUCGAUUGCACCAGCGGACCGCUCUAGCGGUCCAAGACGGCGGCA-----GCACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAU ------.....((.(((......(((((((....))))))).....(((((((-----(...))))))))...)))....)).((((((((((.((((....))))))))))-)))) ( -42.40) >DroSim_CAF1 116116 105 + 1 ------GACUCGCUUGCACCAGCGGACCGCUCUAGCGGUCCAAGGCGGCGGCA-----GCACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAU ------.....((((((....))(((((((....))))))).))))(((((((-----(...)))))))).............((((((((((.((((....))))))))))-)))) ( -44.40) >DroEre_CAF1 116772 87 + 1 ------GACU----UGCACGAGCGGACUUCUCCAGCGGUCC-------------------ACUUGUCGCCACACCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAU ------....----.(((((((.(((((.(....).)))))-------------------.))))).))..............((((((((((.((((....))))))))))-)))) ( -27.60) >DroYak_CAF1 114049 111 + 1 ------GACUCGAUUGCACCAGCGGACCUCUCUAGCGGUCCAAGGCAGCGGCAGUGGGGCACUUGCUGCCACAGCACUAUCCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUUUAU ------.....((((((......(((((.(....).)))))..(((((((..((((...)))))))))))...)))..)))....((((((((.((((....))))))))))))... ( -37.70) >DroPer_CAF1 138263 98 + 1 UCCUGGGGCG----U-GGGGAACAGGCGUCUCUAACGGUCCAAGGCAGCCGCU-----GGAGUC--------GGUGCCAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAU ...((((.((----(-(((((.......))))).))).)))).((((.((((.-----...).)--------)))))).....((((((((((.((((....))))))))))-)))) ( -35.80) >consensus ______GACUCGAUUGCACCAGCGGACCGCUCUAGCGGUCCAAGGCGGCGGCA_____GCACUUGUCGCCACGGCACUAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU_UUAU .....................(((((((........)))))......((((((..........))))))....))........((((((((((.((((....)))))))))).)))) (-15.68 = -17.65 + 1.97)
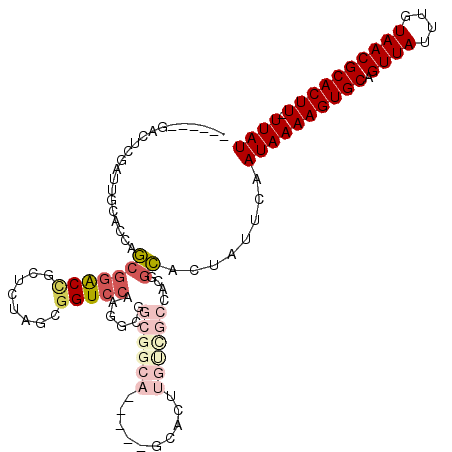
| Location | 2,685,223 – 2,685,328 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -16.15 |
| Energy contribution | -19.37 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2685223 105 - 20766785 AUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUGC-----UGCCGCCGCCUUGGACCGCUAGAGCGGUUCGCUGGUGCAAUCGAGUC------ .(((-((((((((((....)))).)))))))))((((...(..((((((((.((.....-----)).))))))...(((((((....)))))))...))..)..))))...------ ( -42.30) >DroSec_CAF1 117825 105 - 1 AUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUGC-----UGCCGCCGUCUUGGACCGCUAGAGCGGUCCGCUGGUGCAAUCGAGUC------ .(((-((((((((((....)))).)))))))))((((...(..((((((((.(....).-----))))))......(((((((....)))))))...))..)..))))...------ ( -42.70) >DroSim_CAF1 116116 105 - 1 AUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUGC-----UGCCGCCGCCUUGGACCGCUAGAGCGGUCCGCUGGUGCAAGCGAGUC------ ((((-((((((((((....)))).))))))))))......(..((((((((.((.....-----)).))))))...(((((((....)))))))...))..).........------ ( -44.70) >DroEre_CAF1 116772 87 - 1 AUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGGUGUGGCGACAAGU-------------------GGACCGCUGGAGAAGUCCGCUCGUGCA----AGUC------ ((((-((((((((((....)))).))))))))))..............((.((.(((-------------------((((..(....)..))))))).)))).----....------ ( -25.80) >DroYak_CAF1 114049 111 - 1 AUAAAAAGUGCGUUACAAAUAACUGCACUUUUAUUGGAUAGUGCUGUGGCAGCAAGUGCCCCACUGCCGCUGCCUUGGACCGCUAGAGAGGUCCGCUGGUGCAAUCGAGUC------ ...((((((((((((....)))).))))))))(((.(((.(..((((((((((.((((...))))...))))))..(((((........))))))).))..).))).))).------ ( -42.10) >DroPer_CAF1 138263 98 - 1 AUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUGGCACC--------GACUCC-----AGCGGCUGCCUUGGACCGUUAGAGACGCCUGUUCCCC-A----CGCCCCAGGA ((((-((((((((((....)))).)))))))))).....((((((--------(.(...-----.)))).))))((((..(((..(..((....))..)..-)----))..)))).. ( -28.70) >consensus AUAA_AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUAGUGCCGUGGCGACAAGUGC_____UGCCGCCGCCUUGGACCGCUAGAGAGGUCCGCUGGUGCAAUCGAGUC______ ((((.((((((((((....)))).))))))))))......(((((((((((.............))))))......(((((........)))))...)))))............... (-16.15 = -19.37 + 3.22)

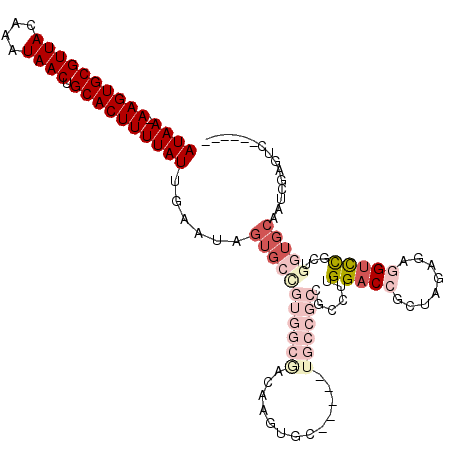
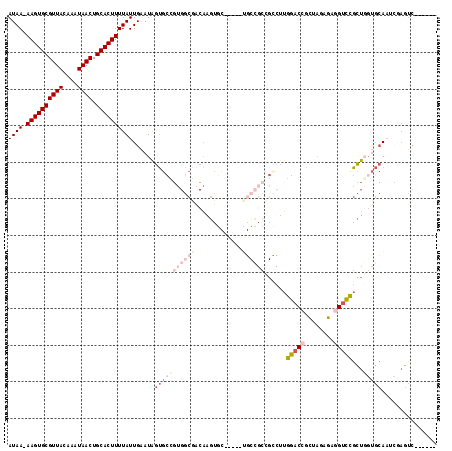
| Location | 2,685,289 – 2,685,388 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2685289 99 + 20766785 UAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAUGGCCACACAUGCGAUGCGAACCCGCACGAUUGGAGCGUUUUGGAUCGGGCGGGGCGGUUC ......((((((((((.((((....))))))))))-))))((((.(...(((...)))..(((((.(((((.(((...))).))))).)))))).)))). ( -33.80) >DroSec_CAF1 117891 99 + 1 UAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAUGGCCACACAUGCGAUGCGAACCCGCACGAUUGGAUCGUUUUGGACCGGGCGGGGCGGUUC ......((((((((((.((((....))))))))))-))))((((.(...(((...)))..(((((.((.((.((.....)).)).)).)))))).)))). ( -30.30) >DroSim_CAF1 116182 99 + 1 UAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU-UUAUGGCCACACAUGCGAUGCGAACCCGCACGAUUGGAUCGUUUUGGACCGGGCGGGGCGGUUC ......((((((((((.((((....))))))))))-))))((((.(...(((...)))..(((((.((.((.((.....)).)).)).)))))).)))). ( -30.30) >DroYak_CAF1 114120 78 + 1 UAUCCAAUAAAAGUGCAGUUAUUUGUAACGCACUUUUUAUGGCCACACAUGCGAUGCGAACCCUCGCGAUUGGAACCG---------------------- ..((((((((((((((.((((....))))))))))))((((......))))...(((((....)))))))))))....---------------------- ( -22.80) >consensus UAUUCAAUAAAAGUGCAGUUAUUUGUAACGCACUU_UUAUGGCCACACAUGCGAUGCGAACCCGCACGAUUGGAUCGUUUUGGACCGGGCGGGGCGGUUC ..((((((..((((((.((((....))))))))))..((((......))))((.((((....)))))))))))).......((((((.......)))))) (-20.27 = -20.40 + 0.13)

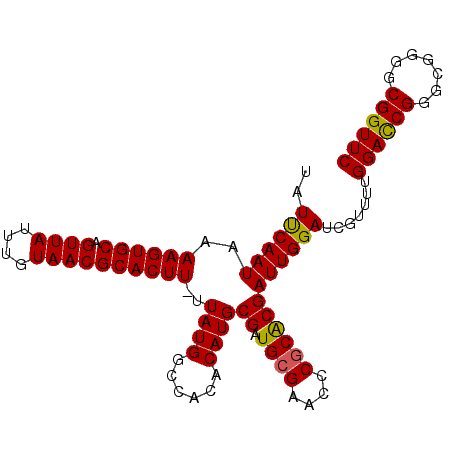
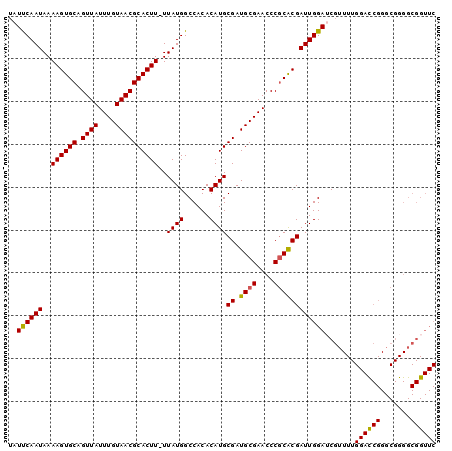
| Location | 2,685,289 – 2,685,388 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2685289 99 - 20766785 GAACCGCCCCGCCCGAUCCAAAACGCUCCAAUCGUGCGGGUUCGCAUCGCAUGUGUGGCCAUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUA ...(((((((((.((((.............)))).)))))...((((...))))))))(.((((-((((((((((....)))).)))))))))).).... ( -29.02) >DroSec_CAF1 117891 99 - 1 GAACCGCCCCGCCCGGUCCAAAACGAUCCAAUCGUGCGGGUUCGCAUCGCAUGUGUGGCCAUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUA ...(((((((((.((((.............)))).)))))...((((...))))))))(.((((-((((((((((....)))).)))))))))).).... ( -28.42) >DroSim_CAF1 116182 99 - 1 GAACCGCCCCGCCCGGUCCAAAACGAUCCAAUCGUGCGGGUUCGCAUCGCAUGUGUGGCCAUAA-AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUA ...(((((((((.((((.............)))).)))))...((((...))))))))(.((((-((((((((((....)))).)))))))))).).... ( -28.42) >DroYak_CAF1 114120 78 - 1 ----------------------CGGUUCCAAUCGCGAGGGUUCGCAUCGCAUGUGUGGCCAUAAAAAGUGCGUUACAAAUAACUGCACUUUUAUUGGAUA ----------------------.(((..((...((((..(....).))))...))..)))...((((((((((((....)))).))))))))........ ( -23.30) >consensus GAACCGCCCCGCCCGGUCCAAAACGAUCCAAUCGUGCGGGUUCGCAUCGCAUGUGUGGCCAUAA_AAGUGCGUUACAAAUAACUGCACUUUUAUUGAAUA ...........................(((..(((((((.......)))))))..))).((....((((((((((....)))).))))))....)).... (-17.85 = -18.35 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:36 2006