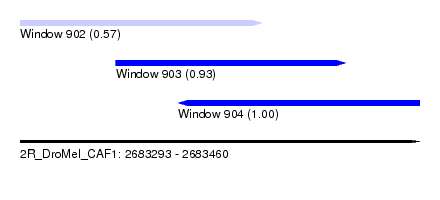
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,683,293 – 2,683,460 |
| Length | 167 |
| Max. P | 0.998862 |

| Location | 2,683,293 – 2,683,394 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -14.83 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.67 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2683293 101 + 20766785 UAUCAAAGAGAAUAUUUCCGUUUGAGGGAUUAAUUAACAACCUGUUAGAACAAGGCAUAAUCUAAUGUGCAUAGUUUACAUCUCAUCUAUCACUAUAAUAU .......((((.((.((((......)))).))..((((.....))))((((...(((((......)))))...))))...))))................. ( -15.90) >DroSec_CAF1 115896 77 + 1 UAUCAAAGAGAUUAUUUCCAUUGGAGGGAUUAAUCAAGAAUCUGUUAGAACAACGCAUAAUCUAAUGUGGAU------------------------AAUAA ((((.....(((((.((((......)))).)))))..................(((((......))))))))------------------------).... ( -14.80) >DroSim_CAF1 114176 77 + 1 UAUCAAAGAGAUUAUUUCCGUUGGAGGGAUUAAUCAACAAUCUAUUAGAACAAGGCAUAAUCUAGUGUGCAU------------------------AAUAA .........(((((.((((......)))).)))))...................(((((......)))))..------------------------..... ( -13.80) >consensus UAUCAAAGAGAUUAUUUCCGUUGGAGGGAUUAAUCAACAAUCUGUUAGAACAAGGCAUAAUCUAAUGUGCAU________________________AAUAA .........(((((.((((......)))).)))))........((((((...........))))))................................... ( -9.99 = -9.67 + -0.33)

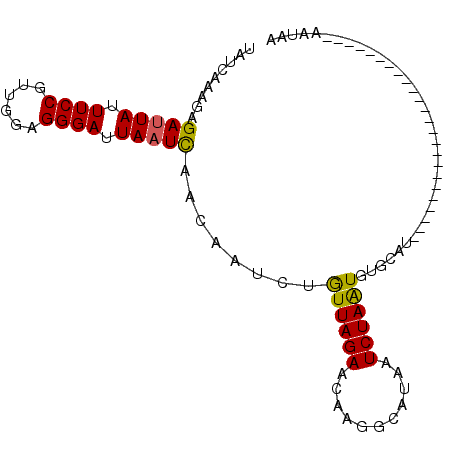
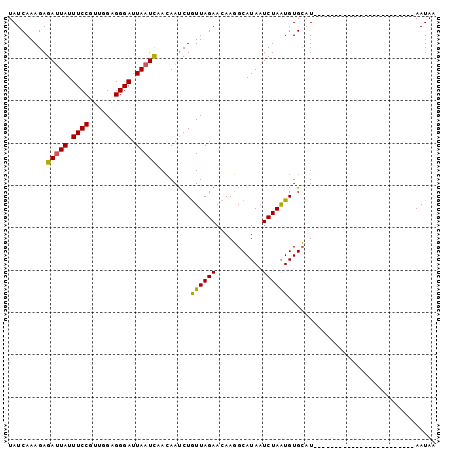
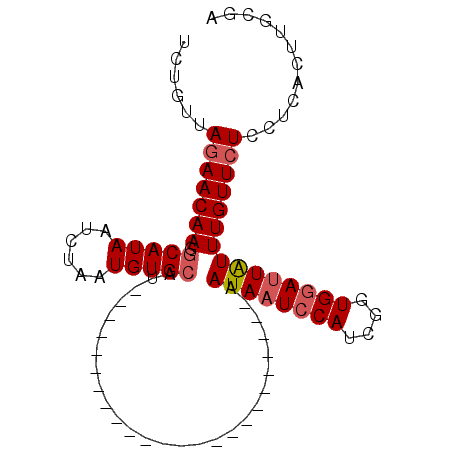
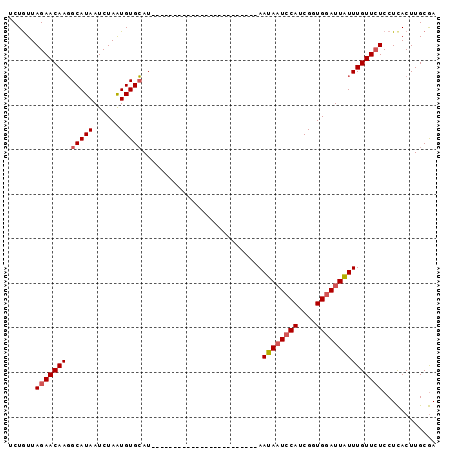
| Location | 2,683,333 – 2,683,429 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.48 |
| Mean single sequence MFE | -16.20 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.87 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2683333 96 + 20766785 CCUGUUAGAACAAGGCAUAAUCUAAUGUGCAUAGUUUACAUCUCAUCUAUCACUAUAAUAUUCCAUCGGUGUAUUGUUUGUUCUCCUUACUUGCAA ......(((((((.(((((......)))))........................(((((((.((...)).))))))))))))))............ ( -14.20) >DroSec_CAF1 115936 72 + 1 UCUGUUAGAACAACGCAUAAUCUAAUGUGGAU------------------------AAUAAUCCAUCGGUGGAUUAUUUGUUCUCCUCACUUGCGA ..((..(((((((...((((((((..((((((------------------------....))))))...)))))))))))))))...))....... ( -18.80) >DroSim_CAF1 114216 72 + 1 UCUAUUAGAACAAGGCAUAAUCUAGUGUGCAU------------------------AAUAAUCCAUCGGUGGAUUAUUUGUUAUCCUCACUUGCGA ..........(((((((((......)))))..------------------------(((((((((....)))))))))...........))))... ( -15.60) >consensus UCUGUUAGAACAAGGCAUAAUCUAAUGUGCAU________________________AAUAAUCCAUCGGUGGAUUAUUUGUUCUCCUCACUUGCGA ......(((((((.(((((......)))))...........................((((((((....)))))))))))))))............ (-10.75 = -11.87 + 1.11)
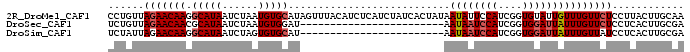
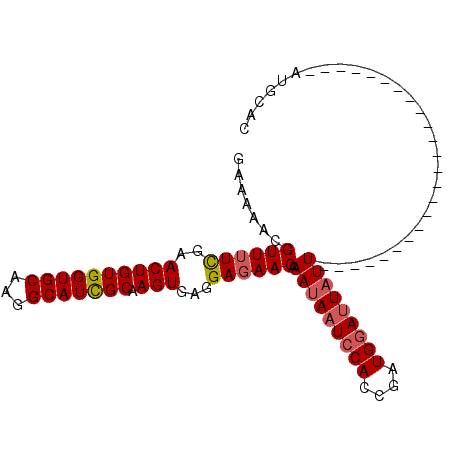
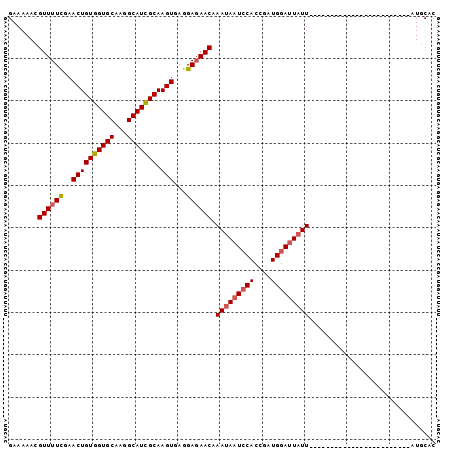
| Location | 2,683,359 – 2,683,460 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2683359 101 - 20766785 GAAAAACGUUUUCGAACUGUGGUGCAAGGCAUUGCAAGUAAGGAGAACAAACAAUACACCGAUGGAAUAUUAUAGUGAUAGAUGAGAUGUAAACUAUGCAC .......((((((..((((..((((...))))..).)))...))))))......................((((((.(((.......)))..))))))... ( -16.40) >DroSec_CAF1 115962 77 - 1 GAAAAACGUUUUCGAACUGUGGUGCAAGGCAUCGCAAGUGAGGAGAACAAAUAAUCCACCGAUGGAUUAUU------------------------AUCCAC .......((((((..((((((((((...))))))).)))...)))))).(((((((((....)))))))))------------------------...... ( -24.80) >DroSim_CAF1 114242 77 - 1 GAAAAACGUUUUUAAACUGUGGUGCAAGGCAUCGCAAGUGAGGAUAACAAAUAAUCCACCGAUGGAUUAUU------------------------AUGCAC .......(((((((...((((((((...))))))))..)))))))....(((((((((....)))))))))------------------------...... ( -20.80) >consensus GAAAAACGUUUUCGAACUGUGGUGCAAGGCAUCGCAAGUGAGGAGAACAAAUAAUCCACCGAUGGAUUAUU________________________AUGCAC .......((((((..((((((((((...))))))).)))...)))))).(((((((((....))))))))).............................. (-17.94 = -18.83 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:33 2006