| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,675,204 – 15,675,309 |
| Length | 105 |
| Max. P | 0.978034 |

| Location | 15,675,204 – 15,675,309 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
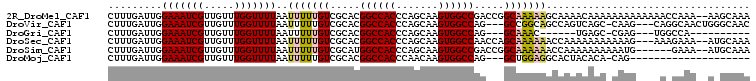
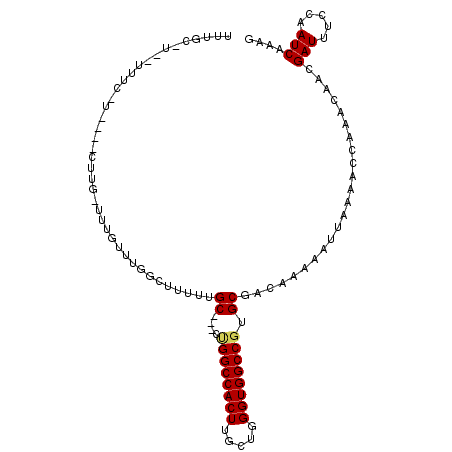
>2R_DroMel_CAF1 15675204 105 + 20766785 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAGCAAGUGGCCGACCGGCAAAAAGCAAAACAAAAAAAAAAAAACCAAA--AAGCAAA ...............(((.(((((((((..(((((((((..(((((((.......)))))))..)))))))))(.....).........)))))))))--.)))... ( -31.50) >DroVir_CAF1 659017 100 + 1 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAGCAAGUGGCCAG---GCCGGCAGCCAGUCAGC-CAAG---CAGGCAACUGGGCAAC ...(((((((((((((.....))))))......(((((((..((((((.......))))))..---).)))))))))))))((-(...---(((....))))))... ( -37.30) >DroGri_CAF1 583628 84 + 1 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAGCAAGUGGCCAG---GCAAAC------UGAGC-CGAG---UGGCCA---------- ...............((..((((((((.....((((((....((((((.......)))))).)---))))).------.))))-))))---..))..---------- ( -27.80) >DroSec_CAF1 552459 102 + 1 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAGCAAGUGGCCAACCAGCAAAAAACCAAAAAAAAAAAG---AAAGAAA--AUGCAAA ......((((((((((.....))))))...(((((((.(...((((((.......))))))...).))))))).))))..........---.......--....... ( -20.60) >DroSim_CAF1 453126 99 + 1 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCAUGGCCACCCAGCAAGUGGCCGACCGGCAAAAAACCAAAAAAAAAAUG------GAAA--AUGCAAA .............(((((.(((((((((.....((((((..(((((((.......)))))))..))))))))))))))).....))))------)...--....... ( -27.40) >DroMoj_CAF1 733937 83 + 1 CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAACAAGUGGCCAG---GCUGGAGGCACUACACA-CAG-------------------- (((((((..(((((((.....))))))).......)))((..((((((.......))))))..---)).))))..........-...-------------------- ( -18.60) >consensus CUUUGAUUGGAAAUCGUUGUUUGGUUUUAAUUUUUGUCGCACGGCCACCCAGCAAGUGGCCAA___GCAAAAAGCCAAACAAA_AAAG____A_GAAA__A_GCAAA .........(((((((.....)))))))..(((((((.....((((((.......)))))).....))))))).................................. (-16.67 = -16.95 + 0.28)
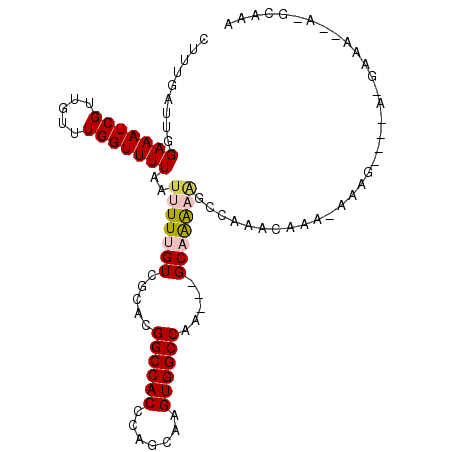
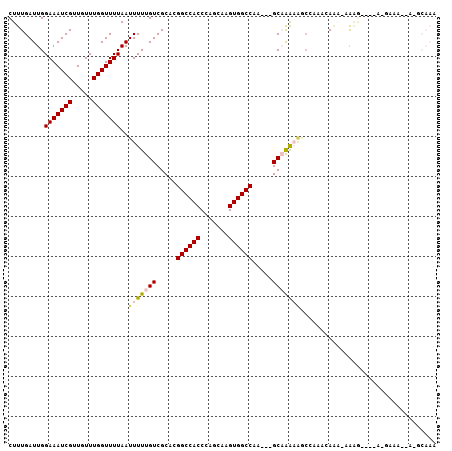
| Location | 15,675,204 – 15,675,309 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15675204 105 - 20766785 UUUGCUU--UUUGGUUUUUUUUUUUUUGUUUUGCUUUUUGCCGGUCGGCCACUUGCUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG .......--(((((((((....((((((((..((.....))..((((((((((.....)))))))).))))))))))..)))))))))....(((.....))).... ( -30.30) >DroVir_CAF1 659017 100 - 1 GUUGCCCAGUUGCCUG---CUUG-GCUGACUGGCUGCCGGC---CUGGCCACUUGCUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG ((((.(((((((((..---...)-)).))))))......((---.((((((((.....)))))))).))...................))))(((.....))).... ( -32.60) >DroGri_CAF1 583628 84 - 1 ----------UGGCCA---CUCG-GCUCA------GUUUGC---CUGGCCACUUGCUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG ----------.(((((---((((-((..(------((..((---...)).))).)))))))))))...........................(((.....))).... ( -23.20) >DroSec_CAF1 552459 102 - 1 UUUGCAU--UUUCUUU---CUUUUUUUUUUUGGUUUUUUGCUGGUUGGCCACUUGCUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG .......--.......---.........(((((((((((((....((((((((.....)))))))).))))........)))))))))....(((.....))).... ( -23.10) >DroSim_CAF1 453126 99 - 1 UUUGCAU--UUUC------CAUUUUUUUUUUGGUUUUUUGCCGGUCGGCCACUUGCUGGGUGGCCAUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG .......--....------.........(((((((((......((((((((((.....)))))))....))).......)))))))))....(((.....))).... ( -22.72) >DroMoj_CAF1 733937 83 - 1 --------------------CUG-UGUGUAGUGCCUCCAGC---CUGGCCACUUGUUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG --------------------..(-(...((((.......((---.((((((((.....)))))))).)).......))))..))........(((.....))).... ( -18.74) >consensus UUUGC_U__UUUC_U____CUUG_UUUGUUUGGCUUUUUGC___CUGGCCACUUGCUGGGUGGCCGUGCGACAAAAAUUAAAACCAAACAACGAUUUCCAAUCAAAG .......................................((....((((((((.....)))))))).)).......................(((.....))).... (-14.56 = -14.62 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:46 2006