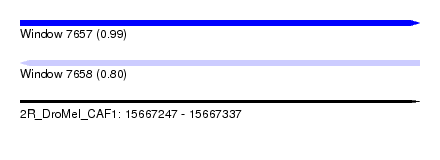
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,667,247 – 15,667,337 |
| Length | 90 |
| Max. P | 0.989084 |

| Location | 15,667,247 – 15,667,337 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15667247 90 + 20766785 -UUUCGUUCCUGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGCCAACAUCAACGGCAACA------GCAACAUCC-ACAUCAACAUCAAGC -...((((..(((((...(((((.((((((.....)))))).)))))...)))))..))))((....------)).......-............... ( -24.00) >DroPse_CAF1 511049 87 + 1 AAGCUGUUCCUGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGCCAACAUCAAC-----------CGGCAACAUCCAACAUCAACAUCAAGC ..(.((((..(((((....((((.((((((.....)))))).))))(((((.........-----------.)))))....)))))..)))).).... ( -24.50) >DroSim_CAF1 445162 90 + 1 -UUCCGUUCCUGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGCCAACAUCAACGGCAACA------GCAACAUCC-ACAUCAACAUCAAGC -..(((((..(((((...(((((.((((((.....)))))).)))))...)))))..))))).....------.........-............... ( -25.40) >DroEre_CAF1 447351 84 + 1 -UUCC-----UGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGC-------AACAGCAACAUUCACAUCCACAUCA-ACAUCAACAUCAAGC -...(-----((((((..(((((.((((((.....)))))).))))).))-------)))))....................-............... ( -23.20) >DroAna_CAF1 434484 69 + 1 -AUCCGAUCCUGUUGUUUAGUUUGAGGCGUUGUCUGUGCCUAAAACUUGGCAACAUCAAC----------------------------AUCAUCAAGC -....(((..((((((..(((((.(((((.......))))).)))))..)))))).....----------------------------)))....... ( -18.40) >DroPer_CAF1 470166 87 + 1 AAGCUGUUCCUGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGCCAACAUCAAC-----------CGGCAACAUCCAACAUCAACAUCAAGC ..(.((((..(((((....((((.((((((.....)))))).))))(((((.........-----------.)))))....)))))..)))).).... ( -24.50) >consensus _AUCCGUUCCUGUUGUUUAGUUUGAGGCGUUGUCUGCGCCUAAAACUUGCCAACAUCAAC_____________GCAACAUCC_ACAUCAACAUCAAGC ..........(((((...(((((.((((((.....)))))).)))))...)))))........................................... (-15.82 = -16.52 + 0.69)

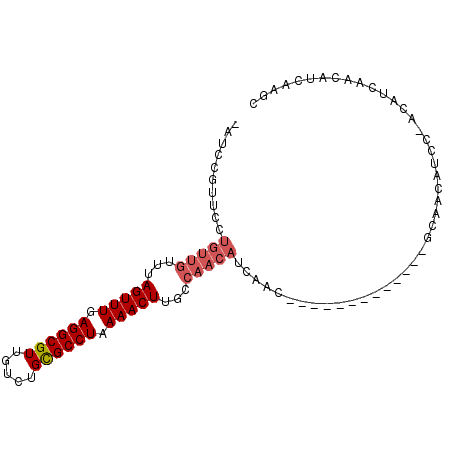
| Location | 15,667,247 – 15,667,337 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.88 |
| Energy contribution | -16.55 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15667247 90 - 20766785 GCUUGAUGUUGAUGU-GGAUGUUGC------UGUUGCCGUUGAUGUUGGCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACAGGAACGAAA- .((((.(((..((((-.((((..((------....)))))).))))..)))(((((.(((((.......))))).)))))......)))).......- ( -26.50) >DroPse_CAF1 511049 87 - 1 GCUUGAUGUUGAUGUUGGAUGUUGCCG-----------GUUGAUGUUGGCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACAGGAACAGCUU ......((((..(((((.....(((((-----------(......))))))(((((.(((((.......))))).)))))...)))))..)))).... ( -26.90) >DroSim_CAF1 445162 90 - 1 GCUUGAUGUUGAUGU-GGAUGUUGC------UGUUGCCGUUGAUGUUGGCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACAGGAACGGAA- ((..((((((.....-.))))))))------.....(((((..(((((...(((((.(((((.......))))).)))))...)))))..)))))..- ( -27.00) >DroEre_CAF1 447351 84 - 1 GCUUGAUGUUGAUGU-UGAUGUGGAUGUGAAUGUUGCUGUU-------GCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACA-----GGAA- ....((((((.((((-(......))))).)))))).(((((-------(..(((((.(((((.......))))).)))))...)))))-----)...- ( -22.90) >DroAna_CAF1 434484 69 - 1 GCUUGAUGAU----------------------------GUUGAUGUUGCCAAGUUUUAGGCACAGACAACGCCUCAAACUAAACAACAGGAUCGGAU- ......((((----------------------------.(((.((((....(((((.((((.........)))).))))).)))).))).))))...- ( -17.40) >DroPer_CAF1 470166 87 - 1 GCUUGAUGUUGAUGUUGGAUGUUGCCG-----------GUUGAUGUUGGCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACAGGAACAGCUU ......((((..(((((.....(((((-----------(......))))))(((((.(((((.......))))).)))))...)))))..)))).... ( -26.90) >consensus GCUUGAUGUUGAUGU_GGAUGUUGC_____________GUUGAUGUUGGCAAGUUUUAGGCGCAGACAACGCCUCAAACUAAACAACAGGAACGGAA_ ......................................(((..(((((...(((((.(((((.......))))).)))))...)))))..)))..... (-14.88 = -16.55 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:41 2006