
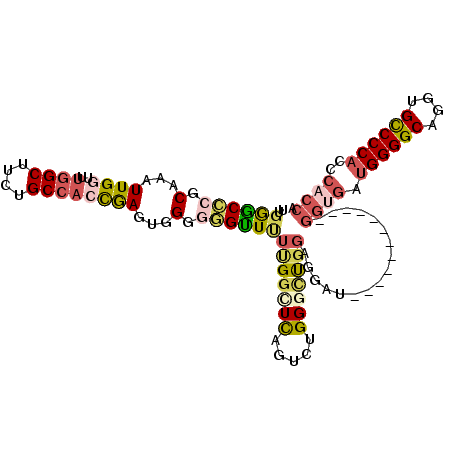
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,614,773 – 15,614,871 |
| Length | 98 |
| Max. P | 0.889219 |

| Location | 15,614,773 – 15,614,871 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -29.21 |
| Energy contribution | -30.47 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15614773 98 + 20766785 UUGGGCCCGCAAAUUGGUUUGGCUUCUGCCACCGAGUGGGGGGUUCUUGGCUCAGUCUGGACUGGAGGAU------------GGUGCUGGGGCAGGUGCCCCACCCACCA .((((.((((...(((((..(((....))))))))))))(((((.((((.(((((((((..(....)..)------------)).)))))).)))).))))).))))... ( -45.20) >DroSec_CAF1 405431 98 + 1 UUGGGCCCGCAAAUUGUUUUGGCUUUUGCCACCGAGUGGGGGGCUUUUGGCUCAGUCUGGACUGGAGGAU------------GGUGUUGGGGCAGGUGCCCCACCCACCA ..((((((.(...(((...((((....)))).)))...).)))))).....(((((....))))).((.(------------((.((.(((((....)))))))))))). ( -40.50) >DroSim_CAF1 388399 98 + 1 UUGGGCCCGCAAAUUGGUUUGGCUUCUGCCACCGAGUGGGGGGCUUUUGGCUCAGUCUGGGCUGGAGGAU------------GGUGAUGGGGCAGGUGCCCCACCCACCA ...(((((.(...(((((..(((....))))))))...).)))))((..((((.....))))..))...(------------((((.((((((....))))))..))))) ( -48.40) >DroEre_CAF1 393256 107 + 1 UUGGGC-CGCAAAUUGGUUUGGC-UCUGCCACCGAGUGGG-GGUUUUUGGCUCGUUCUGGGCCGGGGGAUGGCGAGGGCGAUGGCGAUGGGGCAGGUGCCCCAACCUCCA ((((((-((.....)))).((((-...)))))))).((((-(((..(((.((((((((.(.(((.....)))).))))))).).)))((((((....))))))))))))) ( -45.70) >DroYak_CAF1 409941 110 + 1 UUGGGCCCGCAAAUUGGUUUGGCUUCUGCCACCGAGUGGGGGGUUUUCGGCUUAGUCUGGGUUGGGGGAUAACGAUGGCGAAGGCGAUGGGGCAGGUGCCCCAACCACCA .(((((((.(...(((((..(((....))))))))...).))))...............((((((((.((..(.((.((....)).)).).....)).)))))))).))) ( -45.90) >DroAna_CAF1 380968 89 + 1 UUUAACUCGCAAAUUGGUUUUGCUU-AGCCUGUAAUAGGGAAGUUUGCGUUUUG----GGGCUUGCGU----------------UUUGGGGGCAGAUGUCCCACCCACCG ........(((((.....)))))..-...........((((.((((((.(((..----((((....))----------------))..))))))))).))))........ ( -23.80) >consensus UUGGGCCCGCAAAUUGGUUUGGCUUCUGCCACCGAGUGGGGGGUUUUUGGCUCAGUCUGGGCUGGAGGAU____________GGUGAUGGGGCAGGUGCCCCACCCACCA ..((((((.(...((((..((((....))))))))...).))))))(((((((.....))))))).................((((.((((((....))))))..)))). (-29.21 = -30.47 + 1.26)


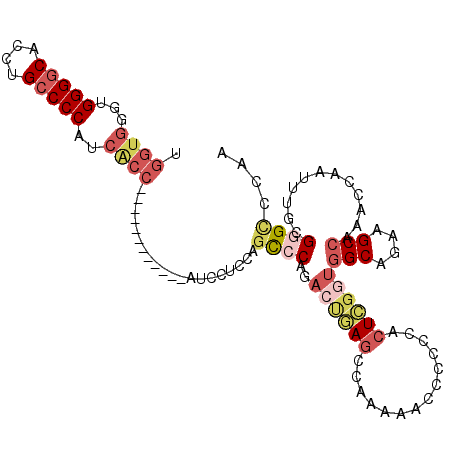
| Location | 15,614,773 – 15,614,871 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -19.61 |
| Energy contribution | -21.34 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15614773 98 - 20766785 UGGUGGGUGGGGCACCUGCCCCAGCACC------------AUCCUCCAGUCCAGACUGAGCCAAGAACCCCCCACUCGGUGGCAGAAGCCAAACCAAUUUGCGGGCCCAA ((((.(.((((((....)))))).))))------------).......((((..((((((..............))))))(((....)))............)))).... ( -33.64) >DroSec_CAF1 405431 98 - 1 UGGUGGGUGGGGCACCUGCCCCAACACC------------AUCCUCCAGUCCAGACUGAGCCAAAAGCCCCCCACUCGGUGGCAAAAGCCAAAACAAUUUGCGGGCCCAA (((((..((((((....)))))).))))------------).....((((....))))........((((.((....))((((....))))...........)))).... ( -36.40) >DroSim_CAF1 388399 98 - 1 UGGUGGGUGGGGCACCUGCCCCAUCACC------------AUCCUCCAGCCCAGACUGAGCCAAAAGCCCCCCACUCGGUGGCAGAAGCCAAACCAAUUUGCGGGCCCAA ((((((.((((((....)))))))))))------------).......((((..((((((..............))))))(((....)))............)))).... ( -38.24) >DroEre_CAF1 393256 107 - 1 UGGAGGUUGGGGCACCUGCCCCAUCGCCAUCGCCCUCGCCAUCCCCCGGCCCAGAACGAGCCAAAAACC-CCCACUCGGUGGCAGA-GCCAAACCAAUUUGCG-GCCCAA .((((((((((((....))))))..)))...((....))..)))...((((((((.((((.........-....)))).((((...-))))......)))).)-)))... ( -35.02) >DroYak_CAF1 409941 110 - 1 UGGUGGUUGGGGCACCUGCCCCAUCGCCUUCGCCAUCGUUAUCCCCCAACCCAGACUAAGCCGAAAACCCCCCACUCGGUGGCAGAAGCCAAACCAAUUUGCGGGCCCAA .((((((.(((((....)))))))))))...............((((((..........(((((...........)))))(((....)))........))).)))..... ( -33.80) >DroAna_CAF1 380968 89 - 1 CGGUGGGUGGGACAUCUGCCCCCAAA----------------ACGCAAGCCC----CAAAACGCAAACUUCCCUAUUACAGGCU-AAGCAAAACCAAUUUGCGAGUUAAA .((.(((..(.....)..)))))...----------------.(((((((..----......))...(((.(((.....)))..-)))..........)))))....... ( -19.30) >consensus UGGUGGGUGGGGCACCUGCCCCAUCACC____________AUCCUCCAGCCCAGACUGAGCCAAAAACCCCCCACUCGGUGGCAGAAGCCAAACCAAUUUGCGGGCCCAA .((((...(((((....)))))..))))....................((((..((((((..............))))))(((....)))............)))).... (-19.61 = -21.34 + 1.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:28 2006