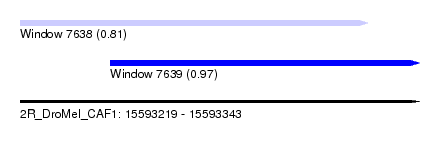
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,593,219 – 15,593,343 |
| Length | 124 |
| Max. P | 0.971175 |

| Location | 15,593,219 – 15,593,327 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
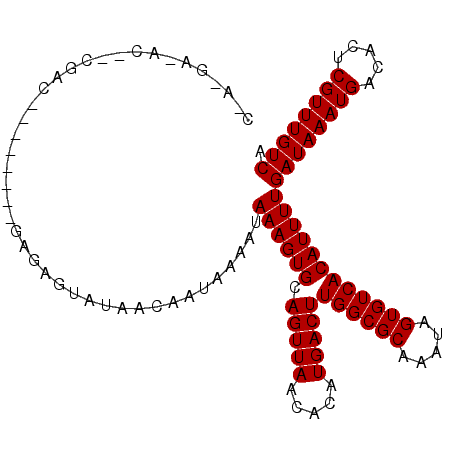
>2R_DroMel_CAF1 15593219 108 + 20766785 CAA-GAUACACCGACAACCCACACAGGGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA ...-(((((.......((((.....))))................(((((((((.....)))))..)))).....)))))......(((((((((.....))))))))) ( -26.30) >DroPse_CAF1 433422 101 + 1 CGA-GAGACU-CGACU------GGAGAGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA ...-...(((-(....------...)))).............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....)))))))). ( -21.10) >DroGri_CAF1 473007 82 + 1 ---------------------------AUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA ---------------------------...............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....)))))))). ( -17.50) >DroMoj_CAF1 597729 83 + 1 C--------------------------AUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA .--------------------------...............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....)))))))). ( -17.50) >DroAna_CAF1 359884 107 + 1 CAAACACACACUCAGAGUCCG--GGGAGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA .....((((((((.(....).--..))))................(((((((((.....)))))..)))).....)))).......(((((((((.....))))))))) ( -21.20) >DroPer_CAF1 392316 101 + 1 CGA-GAGACU-CGACU------GGAGAGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA ...-...(((-(....------...)))).............((((((.(((((.....)))))((((((.....))))))))))))((((((((.....)))))))). ( -21.10) >consensus C_A_GA_AC__CGAC________GAGAGUAUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCA ..........................................((((((.(((((.....)))))((((((.....))))))))))))((((((((.....)))))))). (-17.50 = -17.50 + 0.00)


| Location | 15,593,247 – 15,593,343 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
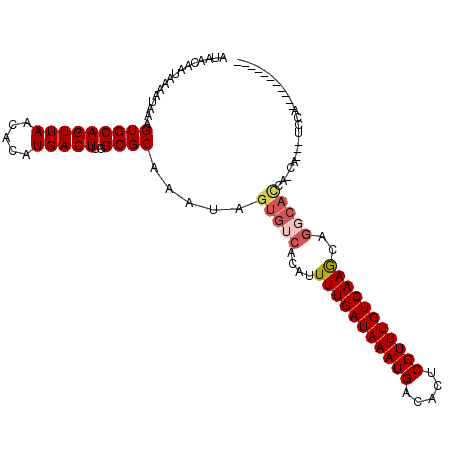
>2R_DroMel_CAF1 15593247 96 + 20766785 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGCACCA-CA---UCC------------ ................(((((((((.....)))))((((((.....))))))...(((((((((((.....)))))))))))...))))..-..---...------------ ( -22.40) >DroPse_CAF1 433443 97 + 1 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAACAGGCACCA-UA---UCCA----------- ................(((((((((.....)))))((((((.....))))))...(((((((((((.....)))))))))))...))))..-..---....----------- ( -22.70) >DroGri_CAF1 473009 98 + 1 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAA----CA-CACGCUCCACA--------- ................(((((((((.....)))))..)))).....((((.....(((((((((((.....)))))))))))...----.)-)))........--------- ( -20.80) >DroSim_CAF1 364126 93 + 1 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGCACCA-CA------------------ ................(((((((((.....)))))((((((.....))))))...(((((((((((.....)))))))))))...))))..-..------------------ ( -22.40) >DroEre_CAF1 371752 99 + 1 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGGCACUCGUUUGUCAAGCAGCCAUCC-CA---UCCACA--------- .................((((((((.....)))))((((((.....))))))....((((((((((.....))))))))))))).......-..---......--------- ( -18.70) >DroWil_CAF1 714003 109 + 1 AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAAUUUGAUAAAUGACACUCGUUUGUCAAACGGGCACCAUCA---UCCAGACUUUGAGGC ....(((.........(((((((((.....)))))..)))).....(((((....(((((((((((.....)))))))))))..))))).....---........))).... ( -24.00) >consensus AUAACAAUAAAAUAAAGUGCAGUUAACACAUGACUUGGCGCAAAUAGUGUCACAUUUUGAUAAAUGACACUCGUUUGUCAAGCAGGCACCA_CA___UCCA___________ ................(((((((((.....)))))..)))).....(((((....(((((((((((.....)))))))))))..)))))....................... (-19.00 = -19.67 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:23 2006