| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,582,485 – 15,582,624 |
| Length | 139 |
| Max. P | 0.963362 |

| Location | 15,582,485 – 15,582,588 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15582485 103 + 20766785 GGAGCAAGUGGAUCGGAGGAUCCGUAAGAUCGGUGGAUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU-UCGGCAUUUCU ((((((((((((((....)))))....((((....))))....)))))..(((((((.((((((((.........)))))))))))))))))-))......... ( -37.70) >DroVir_CAF1 522114 85 + 1 UGUGCAAGUUGCUCGACGGCU-------------------GCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCU ...((..(((....))).))(-------------------((((...((.(((((((.((((((((.........))))))))))))))).)).)))))..... ( -30.00) >DroSim_CAF1 355900 103 + 1 GGAGCAAGUGGAUCGGAGGAUCCGUAAGAUCGGUGGAUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU-UCGGCAUUUCU ((((((((((((((....)))))....((((....))))....)))))..(((((((.((((((((.........)))))))))))))))))-))......... ( -37.70) >DroWil_CAF1 698947 91 + 1 AG--------GCCCGGAGGCAACUGCAACUCGG-----CAGUCGUUUGACAACUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCU ..--------(((.(((((.((.(((......(-----((((.(.....).))))).(((((((((.........)))))))))))).))))))))))...... ( -33.10) >DroAna_CAF1 350666 98 + 1 GGAGCAAGUGGUUCGGUGGCUCGCUGAGG---CUGCCCCGCUUG--UGACAAACGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU-UCGGCAUUUCU (..((((((((...((..(((......))---)..)))))))))--)..)....(((.((((((((.........)))))))))))......-........... ( -38.90) >DroPer_CAF1 383734 102 + 1 GGAGCAAGUUGCUCGACGGCUGCUG-CUGCUGCUCCCGCUGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCC-UCGGCAUUUCU (((((((((((.....)))))((..-..))))))))...(((((...((..((((((.((((((((.........)))))))))))))))).-.)))))..... ( -39.90) >consensus GGAGCAAGUGGCUCGGAGGCUCCGU_AGAUCGGUG___CGGUCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU_UCGGCAUUUCU ........................................((((...((..((((((.((((((((.........))))))))))))))))...))))...... (-21.32 = -21.85 + 0.53)


| Location | 15,582,485 – 15,582,588 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15582485 103 - 20766785 AGAAAUGCCGA-AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAUCCACCGAUCUUACGGAUCCUCCGAUCCACUUGCUCC .........((-..(((((((((.(((((.........))))).)).))))))).)).(((....((((....))))....(((((....)))))....))).. ( -29.60) >DroVir_CAF1 522114 85 - 1 AGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGC-------------------AGCCGUCGAGCAACUUGCACA .((..(((((..(.(((((((((.(((((.........))))).)).))))))).)....))))-------------------)....))..((.....))... ( -29.30) >DroSim_CAF1 355900 103 - 1 AGAAAUGCCGA-AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAUCCACCGAUCUUACGGAUCCUCCGAUCCACUUGCUCC .........((-..(((((((((.(((((.........))))).)).))))))).)).(((....((((....))))....(((((....)))))....))).. ( -29.60) >DroWil_CAF1 698947 91 - 1 AGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAGUUGUCAAACGACUG-----CCGAGUUGCAGUUGCCUCCGGGC--------CU ......(((....((..((((((.(((((.........))))).)).))))....))...((((((-----(......)))))))......)))--------.. ( -29.30) >DroAna_CAF1 350666 98 - 1 AGAAAUGCCGA-AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCGUUUGUCA--CAAGCGGGGCAG---CCUCAGCGAGCCACCGAACCACUUGCUCC ......((.((-..(((((((((.(((((.........))))).)).))))))).)).--....((((((.(---(....))..))).)))........))... ( -30.20) >DroPer_CAF1 383734 102 - 1 AGAAAUGCCGA-GGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGCAGCGGGAGCAGCAG-CAGCAGCCGUCGAGCAACUUGCUCC .(...(((((.-.((((((((((.(((((.........))))).)).))))))..))...))))).).((((((((..-..)).((......))....)))))) ( -37.50) >consensus AGAAAUGCCGA_AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGACCG___CACCGACCU_AAGGAGCCUCCGAGCCACUUGCUCC .........((...(((((((((.(((((.........))))).)).))))))).))............................................... (-17.41 = -17.43 + 0.03)

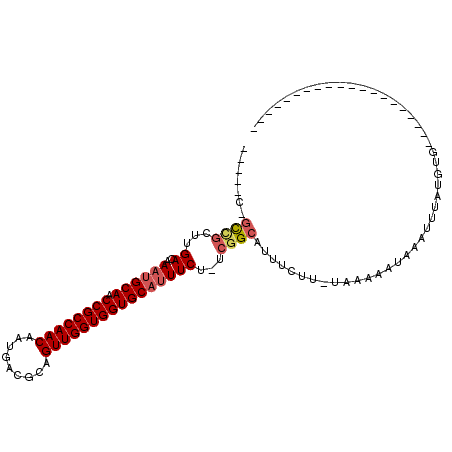
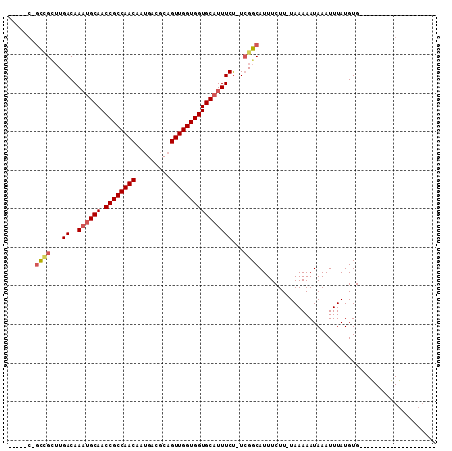
| Location | 15,582,518 – 15,582,624 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15582518 106 + 20766785 UGGAUCGGUGGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU-UCGGCAUUUCUU-UAAAAAUAAAUUUAUGUGGCCGCCUUCGCU-UUGCU-- ..((..((((((((((.(((((((.((((((((.........)))))))))))))))..-)))((((...((-((....))))...))))))))))).))...-.....-- ( -34.50) >DroVir_CAF1 522135 83 + 1 -------GCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCUU-UAAAAAUAAAUUUAUGUG-------------------- -------((((...((.(((((((.((((((((.........))))))))))))))).)).)))).......-..................-------------------- ( -27.40) >DroWil_CAF1 698972 96 + 1 -----CAGUCGUUUGACAACUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCUUUCGGCAUUUCUU-UAAAAAUAAAUUUAUGUGGCCGCCUUUGU--------- -----..(((....)))...((((.((((((((.........)))))))))))).......((((.......-((((......))))....)))).......--------- ( -23.60) >DroMoj_CAF1 585058 84 + 1 -------GCCGCUUGACAAAUGCAACCGCCAACAAAGACGCAGUUGGUGGUGCAUUUCUUUGGGCAUUUCUUUUAAAAAUAAAUUUAUGUG-------------------- -------...(((..(.(((((((.((((((((.........)))))))))))))))..)..)))..........................-------------------- ( -26.40) >DroAna_CAF1 350696 107 + 1 UGCCCCGCUUG--UGACAAACGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU-UCGGCAUUUCUU-UAAAAAUAAAUUUAUGUGCCCGCCGCCGCCUUCGCUGG ...((.((..(--((.(....(((.((((((((.........)))))))))))......-..(((((.....-((((......)))).)))))....).)))....)).)) ( -26.50) >DroPer_CAF1 383766 89 + 1 UCCCGCUGCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCC-UCGGCAUUUCUU-UAAAAAUAAAUUUAUGUG-------------------- ......(((((...((..((((((.((((((((.........)))))))))))))))).-.)))))......-..................-------------------- ( -26.50) >consensus _____C_GCCGCUUGACAAAUGCAACCGCCAACAAUGACGCAGUUGGUGGUGCAUUUCU_UCGGCAUUUCUU_UAAAAAUAAAUUUAUGUG____________________ .......((((...((..((((((.((((((((.........))))))))))))))))...)))).............................................. (-21.93 = -22.40 + 0.47)

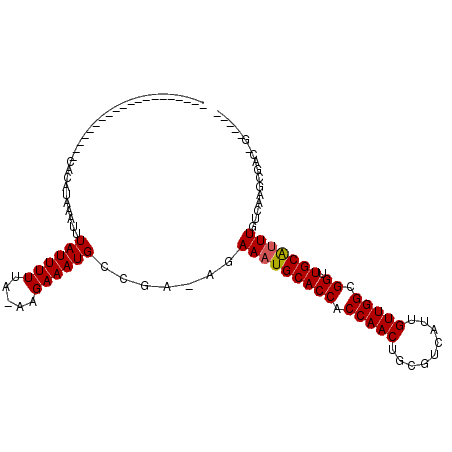
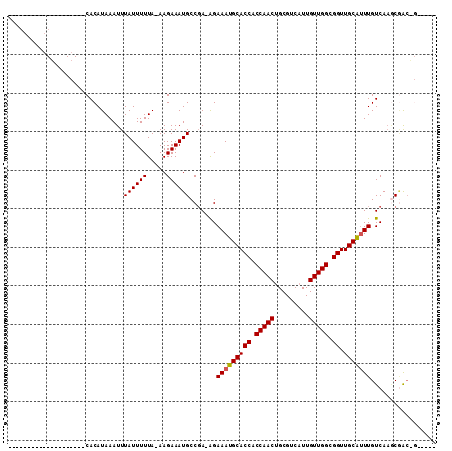
| Location | 15,582,518 – 15,582,624 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15582518 106 - 20766785 --AGCAA-AGCGAAGGCGGCCACAUAAAUUUAUUUUUA-AAGAAAUGCCGA-AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCCACCGAUCCA --.....-..((..((((((........((((....))-)).....)))((-..(((((((((.(((((.........))))).)).))))))).))..)))..))..... ( -28.82) >DroVir_CAF1 522135 83 - 1 --------------------CACAUAAAUUUAUUUUUA-AAGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGC------- --------------------..................-.......((((..(.(((((((((.(((((.........))))).)).))))))).)....))))------- ( -23.60) >DroWil_CAF1 698972 96 - 1 ---------ACAAAGGCGGCCACAUAAAUUUAUUUUUA-AAGAAAUGCCGAAAGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAGUUGUCAAACGACUG----- ---------......((((((..........(((((..-..)))))(((((.....(((((........))))).....)))))))))))(((((.....))))).----- ( -24.80) >DroMoj_CAF1 585058 84 - 1 --------------------CACAUAAAUUUAUUUUUAAAAGAAAUGCCCAAAGAAAUGCACCACCAACUGCGUCUUUGUUGGCGGUUGCAUUUGUCAAGCGGC------- --------------------..........................((((..(.(((((((((.(((((.........))))).)).))))))).)...).)))------- ( -20.30) >DroAna_CAF1 350696 107 - 1 CCAGCGAAGGCGGCGGCGGGCACAUAAAUUUAUUUUUA-AAGAAAUGCCGA-AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCGUUUGUCA--CAAGCGGGGCA ((.((....)).((((((......((((......))))-......))))((-..(((((((((.(((((.........))))).)).))))))).)).--...)))).... ( -32.50) >DroPer_CAF1 383766 89 - 1 --------------------CACAUAAAUUUAUUUUUA-AAGAAAUGCCGA-GGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGGCAGCGGGA --------------------..................-..(...(((((.-.((((((((((.(((((.........))))).)).))))))..))...))))).).... ( -26.20) >consensus ____________________CACAUAAAUUUAUUUUUA_AAGAAAUGCCGA_AGAAAUGCACCACCAACUGCGUCAUUGUUGGCGGUUGCAUUUGUCAAGCGAC_G_____ ..............................((((((.....)))))).......(((((((((.(((((.........))))).)).)))))))................. (-17.27 = -17.30 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:21 2006